Timo Dickscheid
Depth-Wise Representation Development Under Blockwise Self-Supervised Learning for Video Vision Transformers
Jan 14, 2026Abstract:End-to-end backpropagation couples all layers through a global error signal, enabling coordinated learning but requiring long-range credit assignment. Motivated by recent progress in blockwise self-supervised learning (BWSSL), we ask whether masked video transformers can be trained without end-to-end backpropagation. Applying BWSSL to masked video modeling remains relatively underexplored and must handle spatiotemporal context and long-range temporal structure. More broadly, analyses that compare BWSSL and end-to-end training in terms of learning dynamics and depth-wise representation development remain sparse. We apply blockwise learning to a masked autoencoding video vision transformer by partitioning the encoder into blocks, each of which is optimized with a local masked reconstruction loss. Across model sizes and partition granularities, training converges and yields representations close to matched end-to-end baselines under linear-probe and retrieval proxies. In order to compare intermediate representations, we analyze depth-wise decodability, inter-block similarity, and patch-level diagnostics. Blockwise training exposes higher-level structure earlier, while later blocks saturate and operate in a more geometry-preserving regime. It can also induce token-level shifts consistent with stronger early mixing that pooled metrics can miss. These findings point to late-block saturation and interface formation as contributors to the remaining gap.
From Fibers to Cells: Fourier-Based Registration Enables Virtual Cresyl Violet Staining From 3D Polarized Light Imaging
May 16, 2025Abstract:Comprehensive assessment of the various aspects of the brain's microstructure requires the use of complementary imaging techniques. This includes measuring the spatial distribution of cell bodies (cytoarchitecture) and nerve fibers (myeloarchitecture). The gold standard for cytoarchitectonic analysis is light microscopic imaging of cell-body stained tissue sections. To reveal the 3D orientations of nerve fibers, 3D Polarized Light Imaging (3D-PLI) has been introduced as a reliable technique providing a resolution in the micrometer range while allowing processing of series of complete brain sections. 3D-PLI acquisition is label-free and allows subsequent staining of sections after measurement. By post-staining for cell bodies, a direct link between fiber- and cytoarchitecture can potentially be established within the same section. However, inevitable distortions introduced during the staining process make a nonlinear and cross-modal registration necessary in order to study the detailed relationships between cells and fibers in the images. In addition, the complexity of processing histological sections for post-staining only allows for a limited number of samples. In this work, we take advantage of deep learning methods for image-to-image translation to generate a virtual staining of 3D-PLI that is spatially aligned at the cellular level. In a supervised setting, we build on a unique dataset of brain sections, to which Cresyl violet staining has been applied after 3D-PLI measurement. To ensure high correspondence between both modalities, we address the misalignment of training data using Fourier-based registration methods. In this way, registration can be efficiently calculated during training for local image patches of target and predicted staining. We demonstrate that the proposed method enables prediction of a Cresyl violet staining from 3D-PLI, matching individual cell instances.
Analyzing Regional Organization of the Human Hippocampus in 3D-PLI Using Contrastive Learning and Geometric Unfolding
Feb 27, 2024



Abstract:Understanding the cortical organization of the human brain requires interpretable descriptors for distinct structural and functional imaging data. 3D polarized light imaging (3D-PLI) is an imaging modality for visualizing fiber architecture in postmortem brains with high resolution that also captures the presence of cell bodies, for example, to identify hippocampal subfields. The rich texture in 3D-PLI images, however, makes this modality particularly difficult to analyze and best practices for characterizing architectonic patterns still need to be established. In this work, we demonstrate a novel method to analyze the regional organization of the human hippocampus in 3D-PLI by combining recent advances in unfolding methods with deep texture features obtained using a self-supervised contrastive learning approach. We identify clusters in the representations that correspond well with classical descriptions of hippocampal subfields, lending validity to the developed methodology.
Self-Supervised Representation Learning for Nerve Fiber Distribution Patterns in 3D-PLI
Jan 30, 2024Abstract:A comprehensive understanding of the organizational principles in the human brain requires, among other factors, well-quantifiable descriptors of nerve fiber architecture. Three-dimensional polarized light imaging (3D-PLI) is a microscopic imaging technique that enables insights into the fine-grained organization of myelinated nerve fibers with high resolution. Descriptors characterizing the fiber architecture observed in 3D-PLI would enable downstream analysis tasks such as multimodal correlation studies, clustering, and mapping. However, best practices for observer-independent characterization of fiber architecture in 3D-PLI are not yet available. To this end, we propose the application of a fully data-driven approach to characterize nerve fiber architecture in 3D-PLI images using self-supervised representation learning. We introduce a 3D-Context Contrastive Learning (CL-3D) objective that utilizes the spatial neighborhood of texture examples across histological brain sections of a 3D reconstructed volume to sample positive pairs for contrastive learning. We combine this sampling strategy with specifically designed image augmentations to gain robustness to typical variations in 3D-PLI parameter maps. The approach is demonstrated for the 3D reconstructed occipital lobe of a vervet monkey brain. We show that extracted features are highly sensitive to different configurations of nerve fibers, yet robust to variations between consecutive brain sections arising from histological processing. We demonstrate their practical applicability for retrieving clusters of homogeneous fiber architecture and performing data mining for interactively selected templates of specific components of fiber architecture such as U-fibers.
Denoising Diffusion Probabilistic Models for Image Inpainting of Cell Distributions in the Human Brain
Nov 28, 2023



Abstract:Recent advances in imaging and high-performance computing have made it possible to image the entire human brain at the cellular level. This is the basis to study the multi-scale architecture of the brain regarding its subdivision into brain areas and nuclei, cortical layers, columns, and cell clusters down to single cell morphology Methods for brain mapping and cell segmentation exploit such images to enable rapid and automated analysis of cytoarchitecture and cell distribution in complete series of histological sections. However, the presence of inevitable processing artifacts in the image data caused by missing sections, tears in the tissue, or staining variations remains the primary reason for gaps in the resulting image data. To this end we aim to provide a model that can fill in missing information in a reliable way, following the true cell distribution at different scales. Inspired by the recent success in image generation, we propose a denoising diffusion probabilistic model (DDPM), trained on light-microscopic scans of cell-body stained sections. We extend this model with the RePaint method to impute missing or replace corrupted image data. We show that our trained DDPM is able to generate highly realistic image information for this purpose, generating plausible cell statistics and cytoarchitectonic patterns. We validate its outputs using two established downstream task models trained on the same data.
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
GORDA: Graph-based ORientation Distribution Analysis of SLI scatterometry Patterns of Nerve Fibres
Apr 12, 2022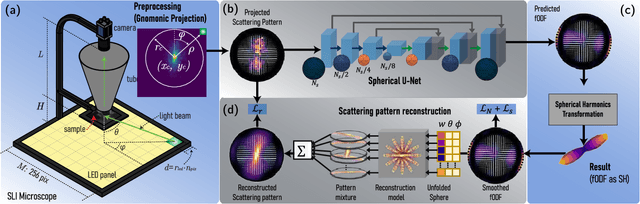
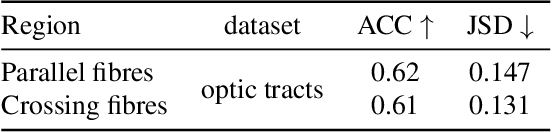
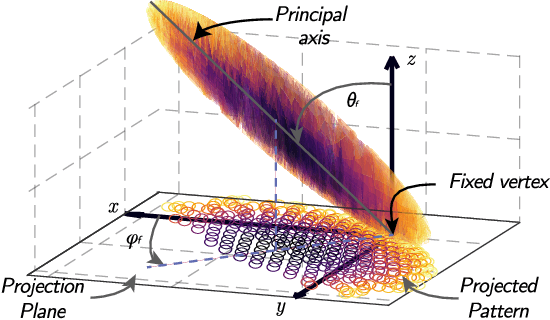
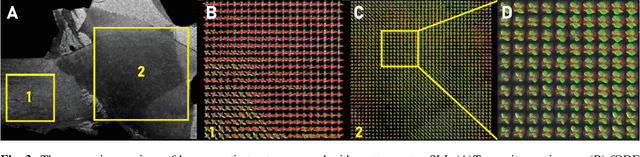
Abstract:Scattered Light Imaging (SLI) is a novel approach for microscopically revealing the fibre architecture of unstained brain sections. The measurements are obtained by illuminating brain sections from different angles and measuring the transmitted (scattered) light under normal incidence. The evaluation of scattering profiles commonly relies on a peak picking technique and feature extraction from the peaks, which allows quantitative determination of parallel and crossing in-plane nerve fibre directions for each image pixel. However, the estimation of the 3D orientation of the fibres cannot be assessed with the traditional methodology. We propose an unsupervised learning approach using spherical convolutions for estimating the 3D orientation of neural fibres, resulting in a more detailed interpretation of the fibre orientation distributions in the brain.
Contour Proposal Networks for Biomedical Instance Segmentation
Apr 07, 2021
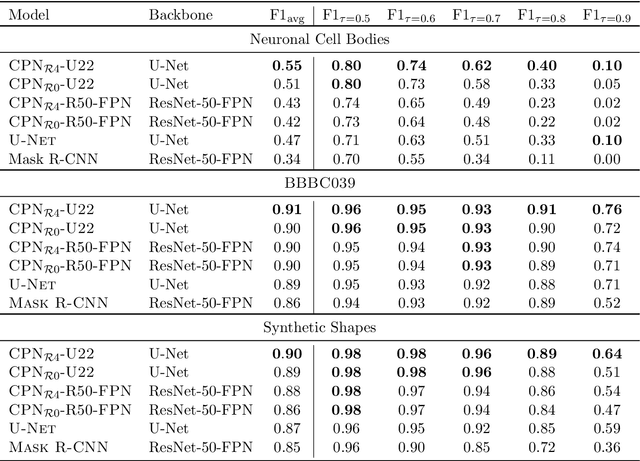
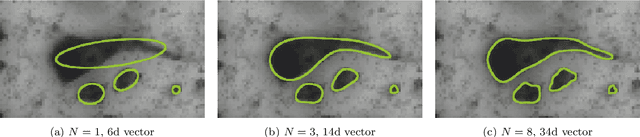
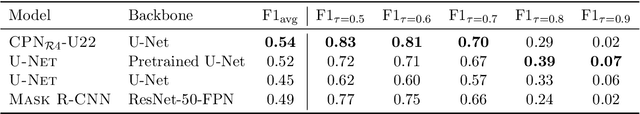
Abstract:We present a conceptually simple framework for object instance segmentation called Contour Proposal Network (CPN), which detects possibly overlapping objects in an image while simultaneously fitting closed object contours using an interpretable, fixed-sized representation based on Fourier Descriptors. The CPN can incorporate state of the art object detection architectures as backbone networks into a single-stage instance segmentation model that can be trained end-to-end. We construct CPN models with different backbone networks, and apply them to instance segmentation of cells in datasets from different modalities. In our experiments, we show CPNs that outperform U-Nets and Mask R-CNNs in instance segmentation accuracy, and present variants with execution times suitable for real-time applications. The trained models generalize well across different domains of cell types. Since the main assumption of the framework are closed object contours, it is applicable to a wide range of detection problems also outside the biomedical domain. An implementation of the model architecture in PyTorch is freely available.
2D histology meets 3D topology: Cytoarchitectonic brain mapping with Graph Neural Networks
Mar 09, 2021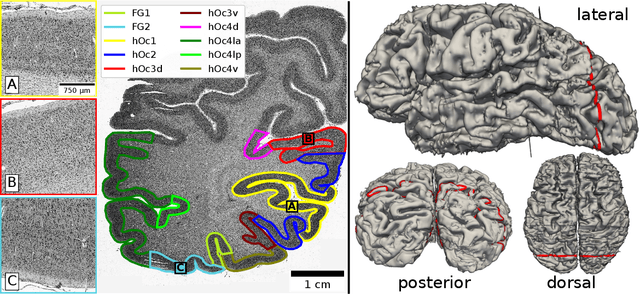
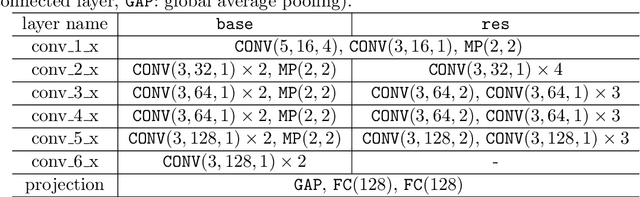
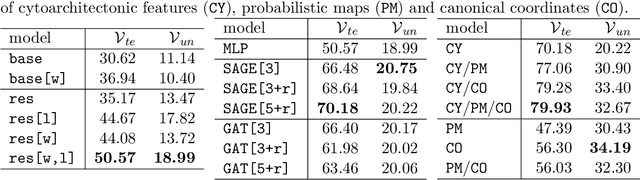
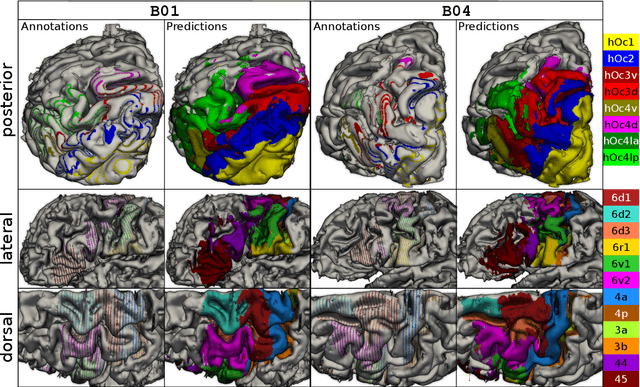
Abstract:Cytoarchitecture describes the spatial organization of neuronal cells in the brain, including their arrangement into layers and columns with respect to cell density, orientation, or presence of certain cell types. It allows to segregate the brain into cortical areas and subcortical nuclei, links structure with connectivity and function, and provides a microstructural reference for human brain atlases. Mapping boundaries between areas requires to scan histological sections at microscopic resolution. While recent high-throughput scanners allow to scan a complete human brain in the order of a year, it is practically impossible to delineate regions at the same pace using the established gold standard method. Researchers have recently addressed cytoarchitectonic mapping of cortical regions with deep neural networks, relying on image patches from individual 2D sections for classification. However, the 3D context, which is needed to disambiguate complex or obliquely cut brain regions, is not taken into account. In this work, we combine 2D histology with 3D topology by reformulating the mapping task as a node classification problem on an approximate 3D midsurface mesh through the isocortex. We extract deep features from cortical patches in 2D histological sections which are descriptive of cytoarchitecture, and assign them to the corresponding nodes on the 3D mesh to construct a large attributed graph. By solving the brain mapping problem on this graph using graph neural networks, we obtain significantly improved classification results. The proposed framework lends itself nicely to integration of additional neuroanatomical priors for mapping.
Contrastive Representation Learning for Whole Brain Cytoarchitectonic Mapping in Histological Human Brain Sections
Nov 25, 2020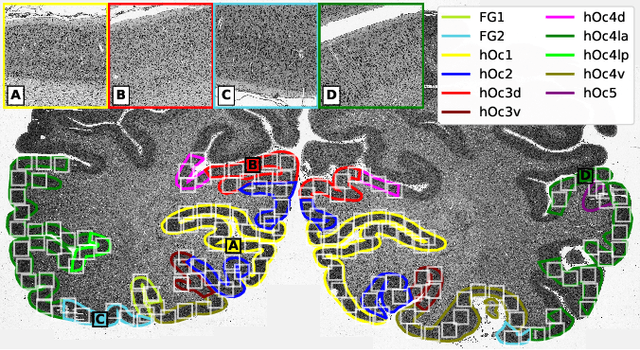
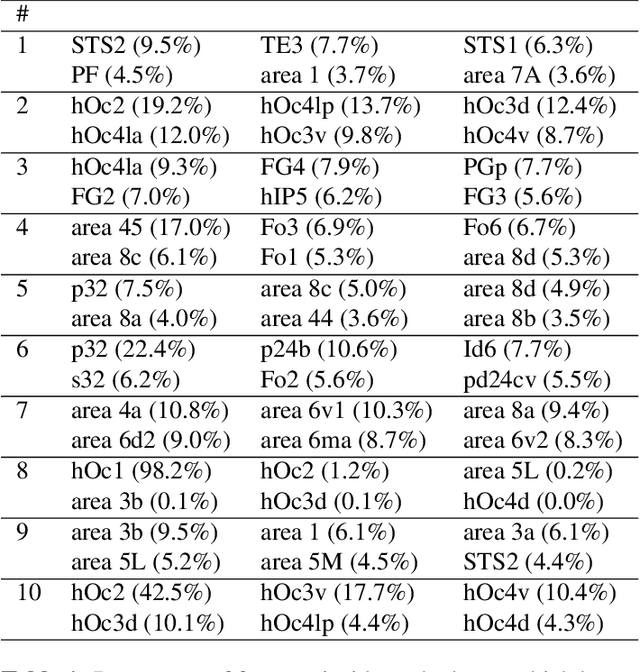
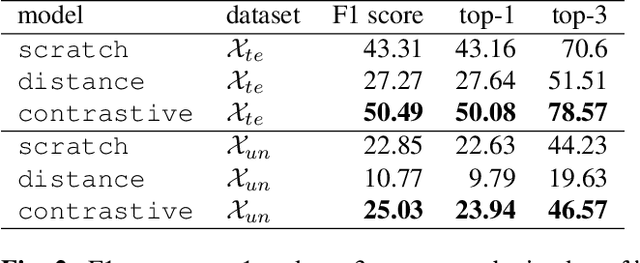
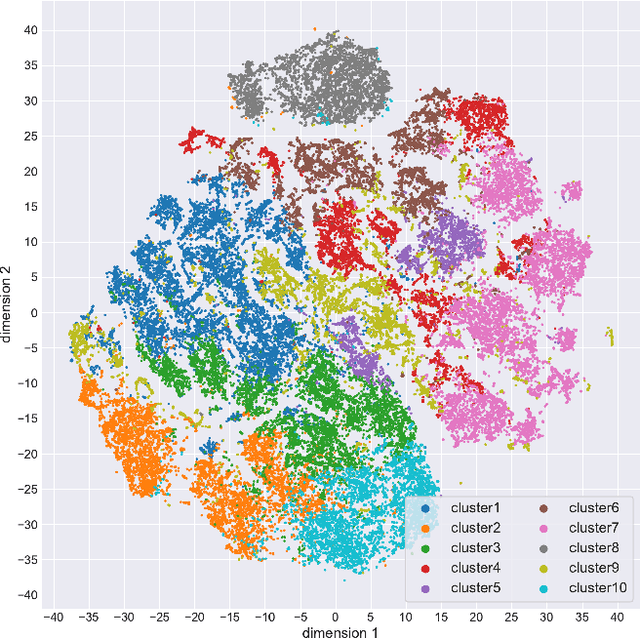
Abstract:Cytoarchitectonic maps provide microstructural reference parcellations of the brain, describing its organization in terms of the spatial arrangement of neuronal cell bodies as measured from histological tissue sections. Recent work provided the first automatic segmentations of cytoarchitectonic areas in the visual system using Convolutional Neural Networks. We aim to extend this approach to become applicable to a wider range of brain areas, envisioning a solution for mapping the complete human brain. Inspired by recent success in image classification, we propose a contrastive learning objective for encoding microscopic image patches into robust microstructural features, which are efficient for cytoarchitectonic area classification. We show that a model pre-trained using this learning task outperforms a model trained from scratch, as well as a model pre-trained on a recently proposed auxiliary task. We perform cluster analysis in the feature space to show that the learned representations form anatomically meaningful groups.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge