Thomas Bocklitz
Denoising and Baseline Correction of Low-Scan FTIR Spectra: A Benchmark of Deep Learning Models Against Traditional Signal Processing
Jan 28, 2026Abstract:High-quality Fourier Transform Infrared (FTIR) imaging usually needs extensive signal averaging to reduce noise and drift which severely limits clinical speed. Deep learning can accelerate imaging by reconstructing spectra from rapid, single-scan inputs. However, separating noise and baseline drift simultaneously without ground truth is an ill-posed inverse problem. Standard black-box architectures often rely on statistical approximations that introduce spectral hallucinations or fail to generalize to unstable atmospheric conditions. To solve these issues we propose a physics-informed cascade Unet that separates denoising and baseline correction tasks using a new, deterministic Physics Bridge. This architecture forces the network to separate random noise from chemical signals using an embedded SNIP layer to enforce spectroscopic constraints instead of learning statistical approximations. We benchmarked this approach against a standard single Unet and a traditional Savitzky-Golay/SNIP workflow. We used a dataset of human hypopharyngeal carcinoma cells (FaDu). The cascade model outperformed all other methods, achieving a 51.3% reduction in RMSE compared to raw single-scan inputs, surpassing both the single Unet (40.2%) and the traditional workflow (33.7%). Peak-aware metrics show that the cascade architecture eliminates spectral hallucinations found in standard deep learning. It also preserves peak intensity with much higher fidelity than traditional smoothing. These results show that the cascade Unet is a robust solution for diagnostic-grade FTIR imaging. It enables imaging speeds 32 times faster than current methods.
Systematic Evaluation of Preprocessing Techniques for Accurate Image Registration in Digital Pathology
Nov 06, 2025Abstract:Image registration refers to the process of spatially aligning two or more images by mapping them into a common coordinate system, so that corresponding anatomical or tissue structures are matched across images. In digital pathology, registration enables direct comparison and integration of information from different stains or imaging modalities, sup-porting applications such as biomarker analysis and tissue reconstruction. Accurate registration of images from different modalities is an essential step in digital pathology. In this study, we investigated how various color transformation techniques affect image registration between hematoxylin and eosin (H&E) stained images and non-linear multimodal images. We used a dataset of 20 tissue sample pairs, with each pair undergoing several preprocessing steps, including different color transformation (CycleGAN, Macenko, Reinhard, Vahadane), inversion, contrast adjustment, intensity normalization, and denoising. All images were registered using the VALIS registration method, which first applies rigid registration and then performs non-rigid registration in two steps on both low and high-resolution images. Registration performance was evaluated using the relative Target Registration Error (rTRE). We reported the median of median rTRE values (MMrTRE) and the average of median rTRE values (AMrTRE) for each method. In addition, we performed a custom point-based evaluation using ten manually selected key points. Registration was done separately for two scenarios, using either the original or inverted multimodal images. In both scenarios, CycleGAN color transformation achieved the lowest registration errors, while the other methods showed higher errors. These findings show that applying color transformation before registration improves alignment between images from different modalities and supports more reliable analysis in digital pathology.
μDeepIQA: deep learning-based fast and robust image quality assessment with local predictions for optical microscopy
Oct 06, 2025



Abstract:Optical microscopy is one of the most widely used techniques in research studies for life sciences and biomedicine. These applications require reliable experimental pipelines to extract valuable knowledge from the measured samples and must be supported by image quality assessment (IQA) to ensure correct processing and analysis of the image data. IQA methods are implemented with variable complexity. However, while most quality metrics have a straightforward implementation, they might be time consuming and computationally expensive when evaluating a large dataset. In addition, quality metrics are often designed for well-defined image features and may be unstable for images out of the ideal domain. To overcome these limitations, recent works have proposed deep learning-based IQA methods, which can provide superior performance, increased generalizability and fast prediction. Our method, named $\mathrm{\mu}$DeepIQA, is inspired by previous studies and applies a deep convolutional neural network designed for IQA on natural images to optical microscopy measurements. We retrained the same architecture to predict individual quality metrics and global quality scores for optical microscopy data. The resulting models provide fast and stable predictions of image quality by generalizing quality estimation even outside the ideal range of standard methods. In addition, $\mathrm{\mu}$DeepIQA provides patch-wise prediction of image quality and can be used to visualize spatially varying quality in a single image. Our study demonstrates that optical microscopy-based studies can benefit from the generalizability of deep learning models due to their stable performance in the presence of outliers, the ability to assess small image patches, and rapid predictions.
GFSR-Net: Guided Focus via Segment-Wise Relevance Network for Interpretable Deep Learning in Medical Imaging
Oct 02, 2025Abstract:Deep learning has achieved remarkable success in medical image analysis, however its adoption in clinical practice is limited by a lack of interpretability. These models often make correct predictions without explaining their reasoning. They may also rely on image regions unrelated to the disease or visual cues, such as annotations, that are not present in real-world conditions. This can reduce trust and increase the risk of misleading diagnoses. We introduce the Guided Focus via Segment-Wise Relevance Network (GFSR-Net), an approach designed to improve interpretability and reliability in medical imaging. GFSR-Net uses a small number of human annotations to approximate where a person would focus within an image intuitively, without requiring precise boundaries or exhaustive markings, making the process fast and practical. During training, the model learns to align its focus with these areas, progressively emphasizing features that carry diagnostic meaning. This guidance works across different types of natural and medical images, including chest X-rays, retinal scans, and dermatological images. Our experiments demonstrate that GFSR achieves comparable or superior accuracy while producing saliency maps that better reflect human expectations. This reduces the reliance on irrelevant patterns and increases confidence in automated diagnostic tools.
ChemRxivQuest: A Curated Chemistry Question-Answer Database Extracted from ChemRxiv Preprints
May 08, 2025Abstract:The rapid expansion of chemistry literature poses significant challenges for researchers seeking to efficiently access domain-specific knowledge. To support advancements in chemistry-focused natural language processing (NLP), we present ChemRxivQuest, a curated dataset of 970 high-quality question-answer (QA) pairs derived from 155 ChemRxiv preprints across 17 subfields of chemistry. Each QA pair is explicitly linked to its source text segment to ensure traceability and contextual accuracy. ChemRxivQuest was constructed using an automated pipeline that combines optical character recognition (OCR), GPT-4o-based QA generation, and a fuzzy matching technique for answer verification. The dataset emphasizes conceptual, mechanistic, applied, and experimental questions, enabling applications in retrieval-based QA systems, search engine development, and fine-tuning of domain-adapted large language models. We analyze the dataset's structure, coverage, and limitations, and outline future directions for expansion and expert validation. ChemRxivQuest provides a foundational resource for chemistry NLP research, education, and tool development.
RAMANMETRIX: a delightful way to analyze Raman spectra
Jan 19, 2022



Abstract:Although Raman spectroscopy is widely used for the investigation of biomedical samples and has a high potential for use in clinical applications, it is not common in clinical routines. One of the factors that obstruct the integration of Raman spectroscopic tools into clinical routines is the complexity of the data processing workflow. Software tools that simplify spectroscopic data handling may facilitate such integration by familiarizing clinical experts with the advantages of Raman spectroscopy. Here, RAMANMETRIX is introduced as a user-friendly software with an intuitive web-based graphical user interface (GUI) that incorporates a complete workflow for chemometric analysis of Raman spectra, from raw data pretreatment to a robust validation of machine learning models. The software can be used both for model training and for the application of the pretrained models onto new data sets. Users have full control of the parameters during model training, but the testing data flow is frozen and does not require additional user input. RAMANMETRIX is available in two versions: as standalone software and web application. Due to the modern software architecture, the computational backend part can be executed separately from the GUI and accessed through an application programming interface (API) for applying a preconstructed model to the measured data. This opens up possibilities for using the software as a data processing backend for the measurement devices in real-time. The models preconstructed by more experienced users can be exported and reused for easy one-click data preprocessing and prediction, which requires minimal interaction between the user and the software. The results of such prediction and graphical outputs of the different data processing steps can be exported and saved.
Sample Size Planning for Classification Models
May 03, 2015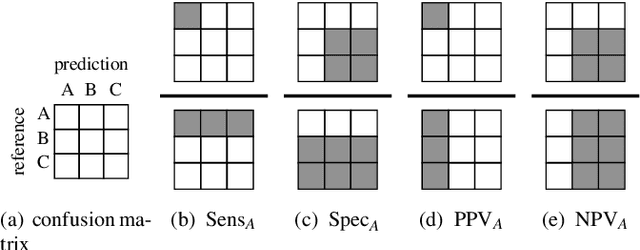
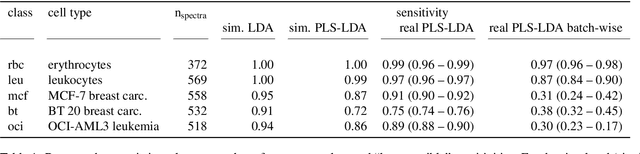
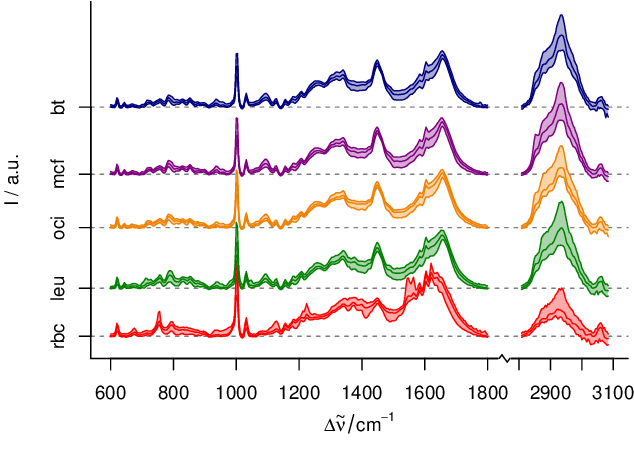
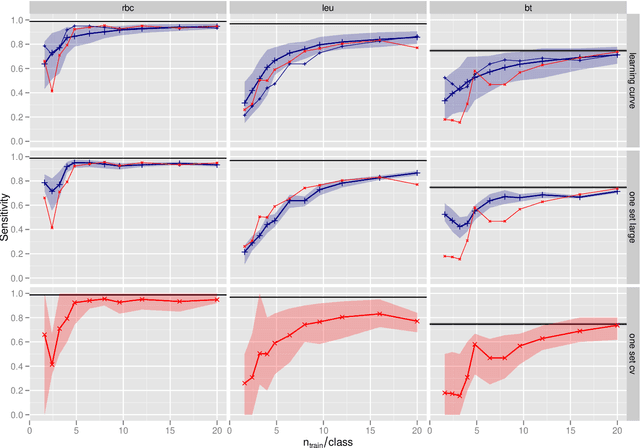
Abstract:In biospectroscopy, suitably annotated and statistically independent samples (e. g. patients, batches, etc.) for classifier training and testing are scarce and costly. Learning curves show the model performance as function of the training sample size and can help to determine the sample size needed to train good classifiers. However, building a good model is actually not enough: the performance must also be proven. We discuss learning curves for typical small sample size situations with 5 - 25 independent samples per class. Although the classification models achieve acceptable performance, the learning curve can be completely masked by the random testing uncertainty due to the equally limited test sample size. In consequence, we determine test sample sizes necessary to achieve reasonable precision in the validation and find that 75 - 100 samples will usually be needed to test a good but not perfect classifier. Such a data set will then allow refined sample size planning on the basis of the achieved performance. We also demonstrate how to calculate necessary sample sizes in order to show the superiority of one classifier over another: this often requires hundreds of statistically independent test samples or is even theoretically impossible. We demonstrate our findings with a data set of ca. 2550 Raman spectra of single cells (five classes: erythrocytes, leukocytes and three tumour cell lines BT-20, MCF-7 and OCI-AML3) as well as by an extensive simulation that allows precise determination of the actual performance of the models in question.
* The paper is published in Analytica Chimica Acta (special issue "CAC2012"). This is a reformatted version of the accepted manuscript with few typos corrected and links to the official publicaion, including the supplementary material (pages 11 - 16 and supplementary-* files in the source). The slides of the presentation at Clircon (2015-04-22, Exeter, UK) are available as ancillary pdf file
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge