Sumanth Nandamuri
SUMNet: Fully Convolutional Model for Fast Segmentation of Anatomical Structures in Ultrasound Volumes
Jan 21, 2019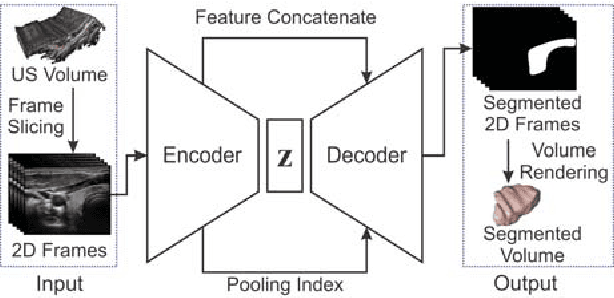
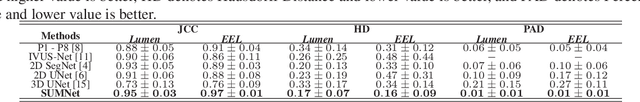
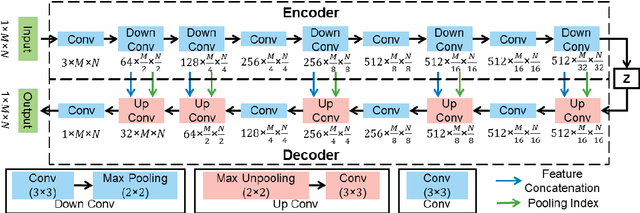
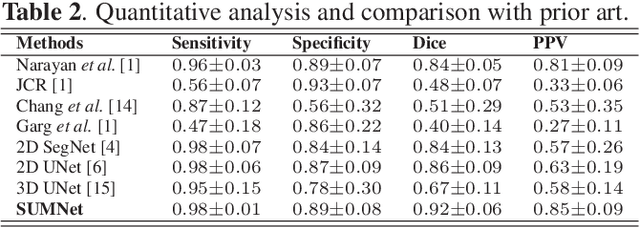
Abstract:Ultrasound imaging is generally employed for real-time investigation of internal anatomy of the human body for disease identification. Delineation of the anatomical boundary of organs and pathological lesions is quite challenging due to the stochastic nature of speckle intensity in the images, which also introduces visual fatigue for the observer. This paper introduces a fully convolutional neural network based method to segment organ and pathologies in ultrasound volume by learning the spatial-relationship between closely related classes in the presence of stochastically varying speckle intensity. We propose a convolutional encoder-decoder like framework with (i) feature concatenation across matched layers in encoder and decoder and (ii) index passing based unpooling at the decoder for semantic segmentation of ultrasound volumes. We have experimentally evaluated the performance on publicly available datasets consisting of $10$ intravascular ultrasound pullback acquired at $20$ MHz and $16$ freehand thyroid ultrasound volumes acquired $11 - 16$ MHz. We have obtained a dice score of $0.93 \pm 0.08$ and $0.92 \pm 0.06$ respectively, following a $10$-fold cross-validation experiment while processing frame of $256 \times 384$ pixel in $0.035$s and a volume of $256 \times 384 \times 384$ voxel in $13.44$s.
UltraCompression: Framework for High Density Compression of Ultrasound Volumes using Physics Modeling Deep Neural Networks
Jan 17, 2019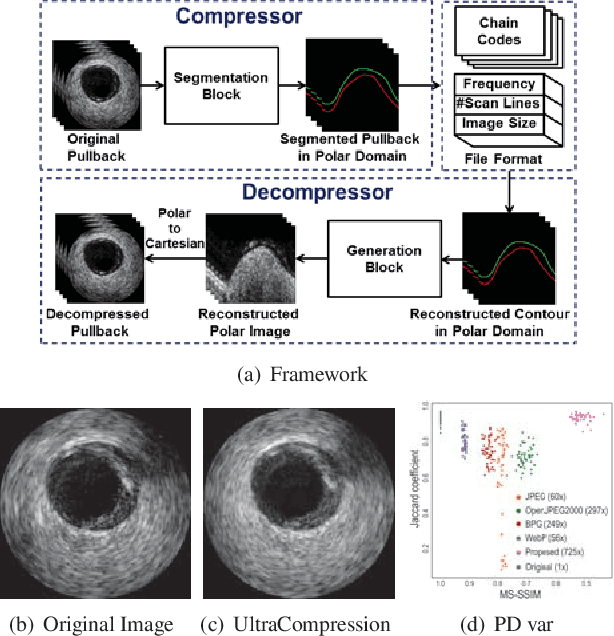
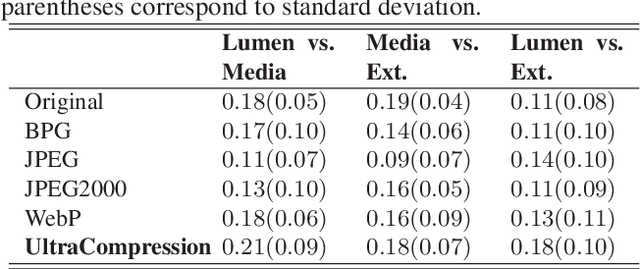
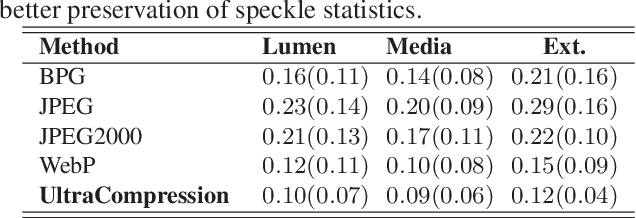
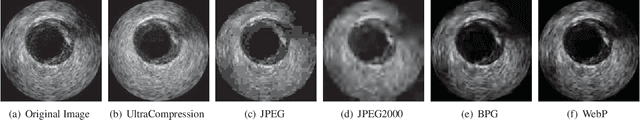
Abstract:Ultrasound image compression by preserving speckle-based key information is a challenging task. In this paper, we introduce an ultrasound image compression framework with the ability to retain realism of speckle appearance despite achieving very high-density compression factors. The compressor employs a tissue segmentation method, transmitting segments along with transducer frequency, number of samples and image size as essential information required for decompression. The decompressor is based on a convolutional network trained to generate patho-realistic ultrasound images which convey essential information pertinent to tissue pathology visible in the images. We demonstrate generalizability of the building blocks using two variants to build the compressor. We have evaluated the quality of decompressed images using distortion losses as well as perception loss and compared it with other off the shelf solutions. The proposed method achieves a compression ratio of $725:1$ while preserving the statistical distribution of speckles. This enables image segmentation on decompressed images to achieve dice score of $0.89 \pm 0.11$, which evidently is not so accurately achievable when images are compressed with current standards like JPEG, JPEG 2000, WebP and BPG. We envision this frame work to serve as a roadmap for speckle image compression standards.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge