Simona Bottani
for The Alzheimer's Disease Neuroimaging Initiative, APPRIMAGE Study Group
Automated MRI Quality Assessment of Brain T1-weighted MRI in Clinical Data Warehouses: A Transfer Learning Approach Relying on Artefact Simulation
Jun 18, 2024



Abstract:The emergence of clinical data warehouses (CDWs), which contain the medical data of millions of patients, has paved the way for vast data sharing for research. The quality of MRIs gathered in CDWs differs greatly from what is observed in research settings and reflects a certain clinical reality. Consequently, a significant proportion of these images turns out to be unusable due to their poor quality. Given the massive volume of MRIs contained in CDWs, the manual rating of image quality is impossible. Thus, it is necessary to develop an automated solution capable of effectively identifying corrupted images in CDWs. This study presents an innovative transfer learning method for automated quality control of 3D gradient echo T1-weighted brain MRIs within a CDW, leveraging artefact simulation. We first intentionally corrupt images from research datasets by inducing poorer contrast, adding noise and introducing motion artefacts. Subsequently, three artefact-specific models are pre-trained using these corrupted images to detect distinct types of artefacts. Finally, the models are generalised to routine clinical data through a transfer learning technique, utilising 3660 manually annotated images. The overall image quality is inferred from the results of the three models, each designed to detect a specific type of artefact. Our method was validated on an independent test set of 385 3D gradient echo T1-weighted MRIs. Our proposed approach achieved excellent results for the detection of bad quality MRIs, with a balanced accuracy of over 87%, surpassing our previous approach by 3.5 percent points. Additionally, we achieved a satisfactory balanced accuracy of 79% for the detection of moderate quality MRIs, outperforming our previous performance by 5 percent points. Our framework provides a valuable tool for exploiting the potential of MRIs in CDWs.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:012
Automatic quality control of brain T1-weighted magnetic resonance images for a clinical data warehouse
Apr 16, 2021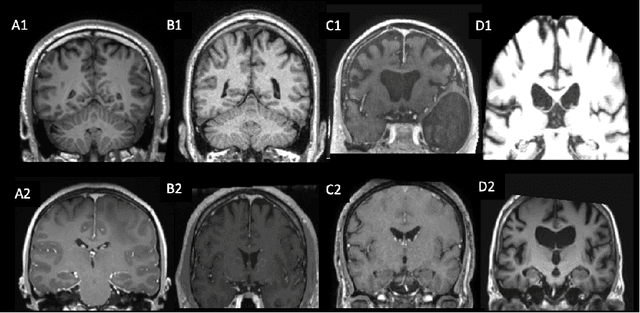
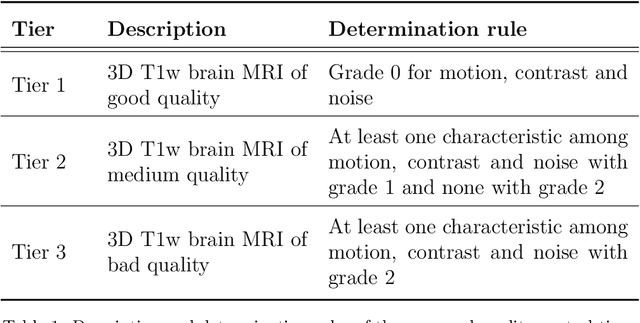
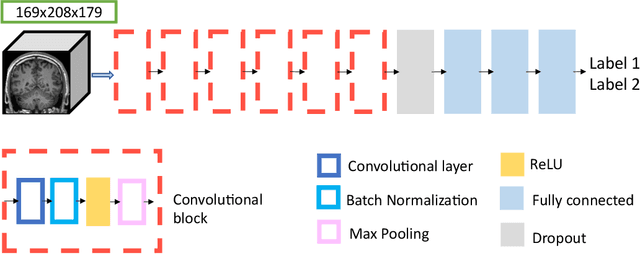
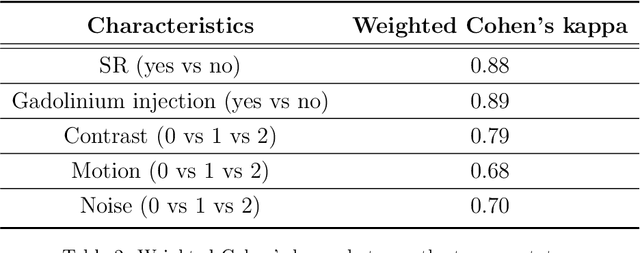
Abstract:Many studies on machine learning (ML) for computer-aided diagnosis have so far been mostly restricted to high-quality research data. Clinical data warehouses, gathering routine examinations from hospitals, offer great promises for training and validation of ML models in a realistic setting. However, the use of such clinical data warehouses requires quality control (QC) tools. Visual QC by experts is time-consuming and does not scale to large datasets. In this paper, we propose a convolutional neural network (CNN) for the automatic QC of 3D T1-weighted brain MRI for a large heterogeneous clinical data warehouse. To that purpose, we used the data warehouse of the hospitals of the Greater Paris area (Assistance Publique-H\^opitaux de Paris [AP-HP]). Specifically, the objectives were: 1) to identify images which are not proper T1-weighted brain MRIs; 2) to identify acquisitions for which gadolinium was injected; 3) to rate the overall image quality. We used 5000 images for training and validation and a separate set of 500 images for testing. In order to train/validate the CNN, the data were annotated by two trained raters according to a visual QC protocol that we specifically designed for application in the setting of a data warehouse. For objectives 1 and 2, our approach achieved excellent accuracy (balanced accuracy and F1-score \textgreater 90\%), similar to the human raters. For objective 3, the performance was good but substantially lower than that of human raters. Nevertheless, the automatic approach accurately identified (balanced accuracy and F1-score \textgreater 80\%) low quality images, which would typically need to be excluded. Overall, our approach shall be useful for exploiting hospital data warehouses in medical image computing.
Gaussian Graphical Model exploration and selection in high dimension low sample size setting
Mar 11, 2020
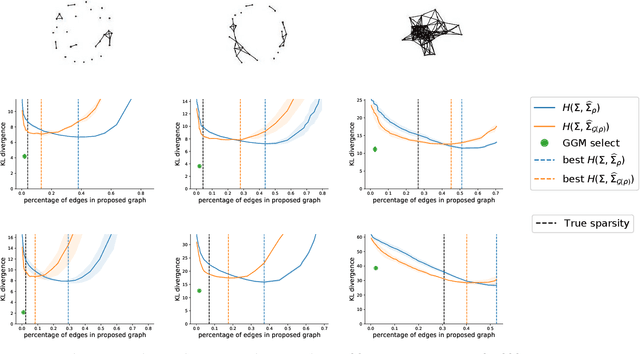
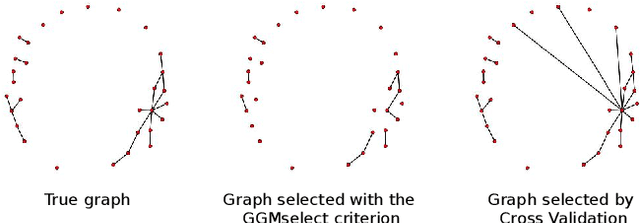
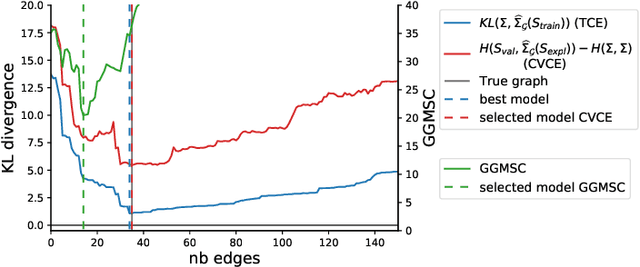
Abstract:Gaussian Graphical Models (GGM) are often used to describe the conditional correlations between the components of a random vector. In this article, we compare two families of GGM inference methods: nodewise edge selection and penalised likelihood maximisation. We demonstrate on synthetic data that, when the sample size is small, the two methods produce graphs with either too few or too many edges when compared to the real one. As a result, we propose a composite procedure that explores a family of graphs with an nodewise numerical scheme and selects a candidate among them with an overall likelihood criterion. We demonstrate that, when the number of observations is small, this selection method yields graphs closer to the truth and corresponding to distributions with better KL divergence with regards to the real distribution than the other two. Finally, we show the interest of our algorithm on two concrete cases: first on brain imaging data, then on biological nephrology data. In both cases our results are more in line with current knowledge in each field.
Convolutional Neural Networks for Classification of Alzheimer's Disease: Overview and Reproducible Evaluation
Apr 16, 2019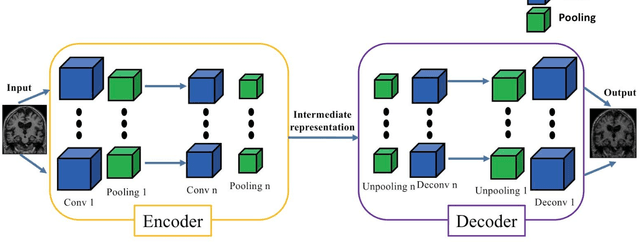
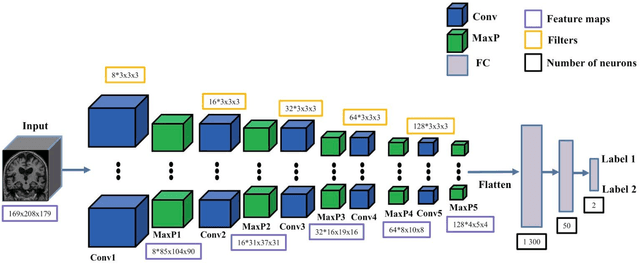
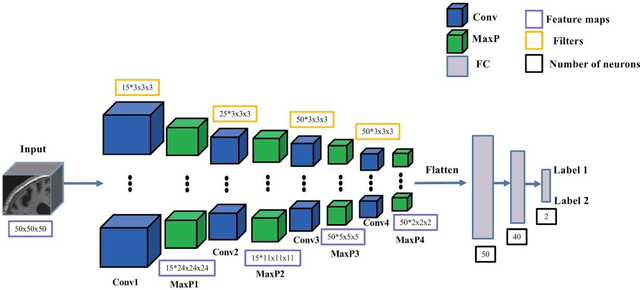
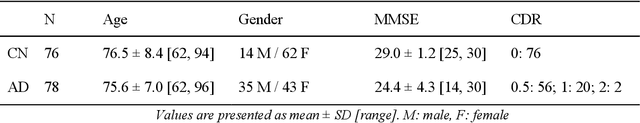
Abstract:In the past two years, over 30 papers have proposed to use convolutional neural network (CNN) for AD classification. However, the classification performances across studies are difficult to compare. Moreover, these studies are hardly reproducible because their frameworks are not publicly accessible. Lastly, some of these papers may reported biased performances due to inadequate or unclear validation procedure and also it is unclear how the model architecture and parameters were chosen. In the present work, we aim to address these limitations through three main contributions. First, we performed a systematic literature review of studies using CNN for AD classification from anatomical MRI. We identified four main types of approaches: 2D slice-level, 3D patch-level, ROI-based and 3D subject-level CNN. Moreover, we found that more than half of the surveyed papers may have suffered from data leakage and thus reported biased performances. Our second contribution is an open-source framework for classification of AD. Thirdly, we used this framework to rigorously compare different CNN architectures, which are representative of the existing literature, and to study the influence of key components on classification performances. On the validation set, the ROI-based (hippocampus) CNN achieved highest balanced accuracy (0.86 for AD vs CN and 0.80 for sMCI vs pMCI) compared to other approaches. Transfer learning with autoencoder pre-training did not improve the average accuracy but reduced the variance. Training using longitudinal data resulted in similar or higher performance, depending on the approach, compared to training with only baseline data. Sophisticated image preprocessing did not improve the results. Lastly, CNN performed similarly to standard SVM for task AD vs CN but outperformed SVM for task sMCI vs pMCI, demonstrating the potential of deep learning for challenging diagnostic tasks.
Reproducible evaluation of diffusion MRI features for automatic classification of patients with Alzheimers disease
Dec 28, 2018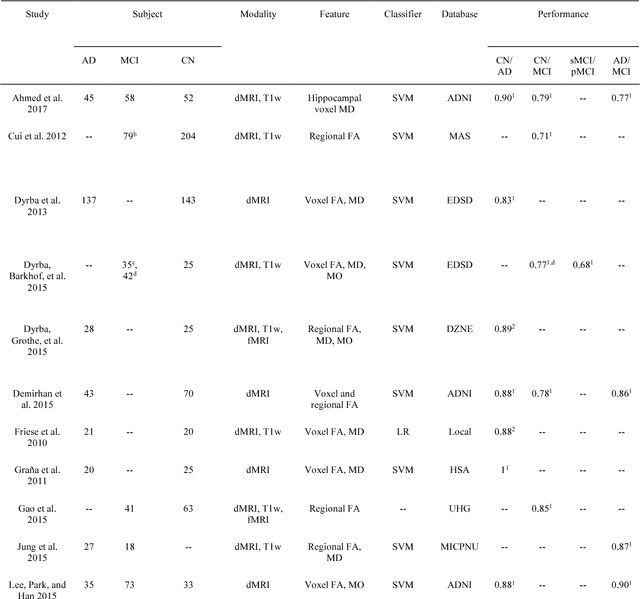
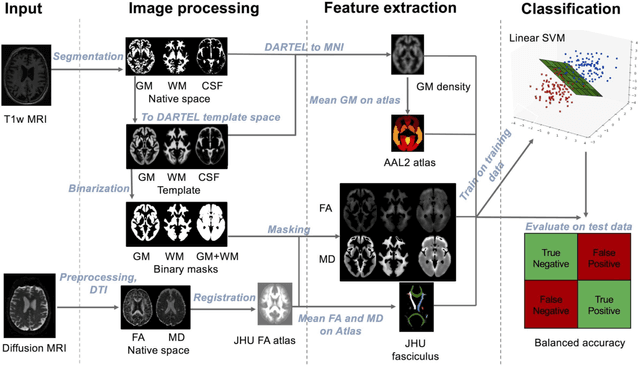
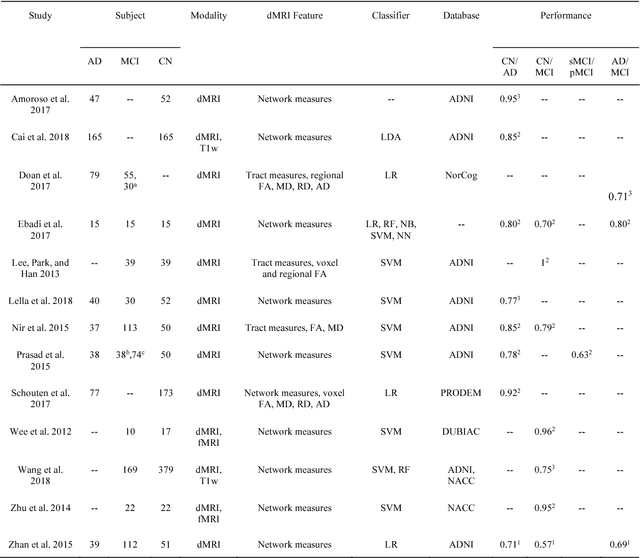
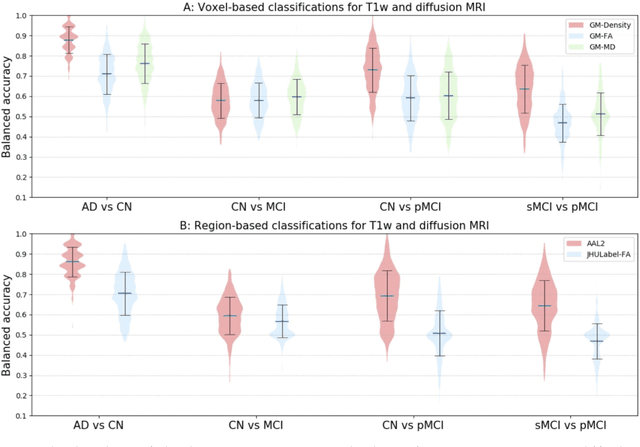
Abstract:Diffusion MRI is the modality of choice to study alterations of white matter. In the past years, various works have used diffusion MRI for automatic classification of Alzheimers disease. However, the performances obtained with different approaches are difficult to compare because of variations in components such as input data, participant selection, image preprocessing, feature extraction, feature selection (FS) and cross-validation (CV) procedure. Moreover, these studies are also difficult to reproduce because these different components are not readily available. In a previous work (Samper-Gonzalez et al. 2018), we proposed an open-source framework for the reproducible evaluation of AD classification from T1-weighted (T1w) MRI and PET data. In the present paper, we extend this framework to diffusion MRI data. The framework comprises: tools to automatically convert ADNI data into the BIDS standard, pipelines for image preprocessing and feature extraction, baseline classifiers and a rigorous CV procedure. We demonstrate the use of the framework through assessing the influence of diffusion tensor imaging (DTI) metrics (fractional anisotropy - FA, mean diffusivity - MD), feature types, imaging modalities (diffusion MRI or T1w MRI), data imbalance and FS bias. First, voxel-wise features generally gave better performances than regional features. Secondly, FA and MD provided comparable results for voxel-wise features. Thirdly, T1w MRI performed better than diffusion MRI. Fourthly, we demonstrated that using non-nested validation of FS leads to unreliable and over-optimistic results. All the code is publicly available: general-purpose tools have been integrated into the Clinica software (www.clinica.run) and the paper-specific code is available at: https://gitlab.icm-institute.org/aramislab/AD-ML.
Reproducible evaluation of classification methods in Alzheimer's disease: framework and application to MRI and PET data
Aug 20, 2018
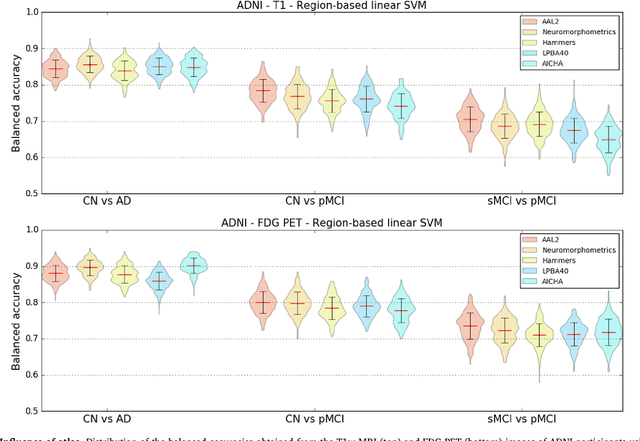
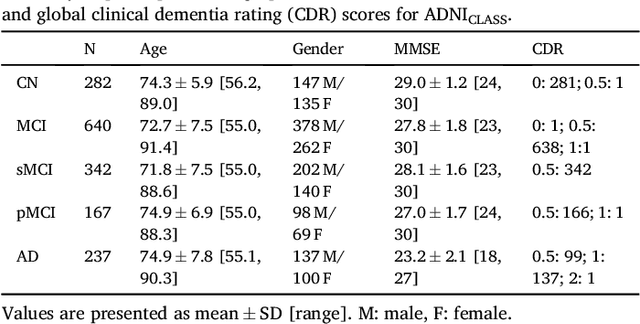
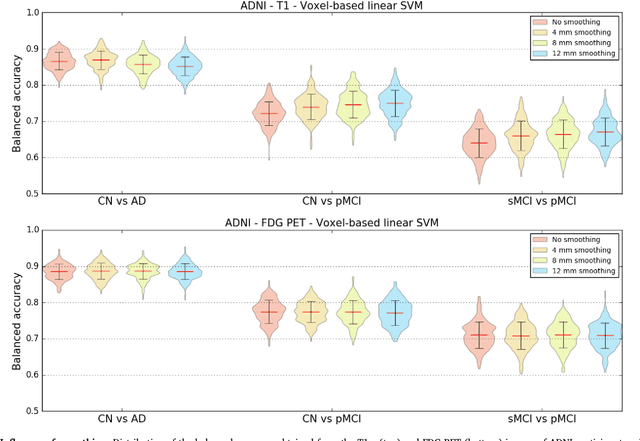
Abstract:A large number of papers have introduced novel machine learning and feature extraction methods for automatic classification of AD. However, they are difficult to reproduce because key components of the validation are often not readily available. These components include selected participants and input data, image preprocessing and cross-validation procedures. The performance of the different approaches is also difficult to compare objectively. In particular, it is often difficult to assess which part of the method provides a real improvement, if any. We propose a framework for reproducible and objective classification experiments in AD using three publicly available datasets (ADNI, AIBL and OASIS). The framework comprises: i) automatic conversion of the three datasets into BIDS format, ii) a modular set of preprocessing pipelines, feature extraction and classification methods, together with an evaluation framework, that provide a baseline for benchmarking the different components. We demonstrate the use of the framework for a large-scale evaluation on 1960 participants using T1 MRI and FDG PET data. In this evaluation, we assess the influence of different modalities, preprocessing, feature types, classifiers, training set sizes and datasets. Performances were in line with the state-of-the-art. FDG PET outperformed T1 MRI for all classification tasks. No difference in performance was found for the use of different atlases, image smoothing, partial volume correction of FDG PET images, or feature type. Linear SVM and L2-logistic regression resulted in similar performance and both outperformed random forests. The classification performance increased along with the number of subjects used for training. Classifiers trained on ADNI generalized well to AIBL and OASIS. All the code of the framework and the experiments is publicly available at: https://gitlab.icm-institute.org/aramislab/AD-ML.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge