Shuo Zhao
Ocean University of China
An Active Learning Pipeline for Biomedical Image Instance Segmentation with Minimal Human Intervention
Nov 06, 2025Abstract:Biomedical image segmentation is critical for precise structure delineation and downstream analysis. Traditional methods often struggle with noisy data, while deep learning models such as U-Net have set new benchmarks in segmentation performance. nnU-Net further automates model configuration, making it adaptable across datasets without extensive tuning. However, it requires a substantial amount of annotated data for cross-validation, posing a challenge when only raw images but no labels are available. Large foundation models offer zero-shot generalizability, but may underperform on specific datasets with unique characteristics, limiting their direct use for analysis. This work addresses these bottlenecks by proposing a data-centric AI workflow that leverages active learning and pseudo-labeling to combine the strengths of traditional neural networks and large foundation models while minimizing human intervention. The pipeline starts by generating pseudo-labels from a foundation model, which are then used for nnU-Net's self-configuration. Subsequently, a representative core-set is selected for minimal manual annotation, enabling effective fine-tuning of the nnU-Net model. This approach significantly reduces the need for manual annotations while maintaining competitive performance, providing an accessible solution for biomedical researchers to apply state-of-the-art AI techniques in their segmentation tasks. The code is available at https://github.com/MMV-Lab/AL_BioMed_img_seg.
* 6 pages, 4 figures, presented at Bildverarbeitung f\"ur die Medizin (BVM) 2025, Wiesbaden, Germany
Data Efficiency and Transfer Robustness in Biomedical Image Segmentation: A Study of Redundancy and Forgetting with Cellpose
Nov 06, 2025Abstract:Generalist biomedical image segmentation models such as Cellpose are increasingly applied across diverse imaging modalities and cell types. However, two critical challenges remain underexplored: (1) the extent of training data redundancy and (2) the impact of cross domain transfer on model retention. In this study, we conduct a systematic empirical analysis of these challenges using Cellpose as a case study. First, to assess data redundancy, we propose a simple dataset quantization (DQ) strategy for constructing compact yet diverse training subsets. Experiments on the Cyto dataset show that image segmentation performance saturates with only 10% of the data, revealing substantial redundancy and potential for training with minimal annotations. Latent space analysis using MAE embeddings and t-SNE confirms that DQ selected patches capture greater feature diversity than random sampling. Second, to examine catastrophic forgetting, we perform cross domain finetuning experiments and observe significant degradation in source domain performance, particularly when adapting from generalist to specialist domains. We demonstrate that selective DQ based replay reintroducing just 5-10% of the source data effectively restores source performance, while full replay can hinder target adaptation. Additionally, we find that training domain sequencing improves generalization and reduces forgetting in multi stage transfer. Our findings highlight the importance of data centric design in biomedical image segmentation and suggest that efficient training requires not only compact subsets but also retention aware learning strategies and informed domain ordering. The code is available at https://github.com/MMV-Lab/biomedseg-efficiency.
PathMR: Multimodal Visual Reasoning for Interpretable Pathology Diagnosis
Aug 28, 2025Abstract:Deep learning based automated pathological diagnosis has markedly improved diagnostic efficiency and reduced variability between observers, yet its clinical adoption remains limited by opaque model decisions and a lack of traceable rationale. To address this, recent multimodal visual reasoning architectures provide a unified framework that generates segmentation masks at the pixel level alongside semantically aligned textual explanations. By localizing lesion regions and producing expert style diagnostic narratives, these models deliver the transparent and interpretable insights necessary for dependable AI assisted pathology. Building on these advancements, we propose PathMR, a cell-level Multimodal visual Reasoning framework for Pathological image analysis. Given a pathological image and a textual query, PathMR generates expert-level diagnostic explanations while simultaneously predicting cell distribution patterns. To benchmark its performance, we evaluated our approach on the publicly available PathGen dataset as well as on our newly developed GADVR dataset. Extensive experiments on these two datasets demonstrate that PathMR consistently outperforms state-of-the-art visual reasoning methods in text generation quality, segmentation accuracy, and cross-modal alignment. These results highlight the potential of PathMR for improving interpretability in AI-driven pathological diagnosis. The code will be publicly available in https://github.com/zhangye-zoe/PathMR.
IPA-NeRF: Illusory Poisoning Attack Against Neural Radiance Fields
Jul 16, 2024



Abstract:Neural Radiance Field (NeRF) represents a significant advancement in computer vision, offering implicit neural network-based scene representation and novel view synthesis capabilities. Its applications span diverse fields including robotics, urban mapping, autonomous navigation, virtual reality/augmented reality, etc., some of which are considered high-risk AI applications. However, despite its widespread adoption, the robustness and security of NeRF remain largely unexplored. In this study, we contribute to this area by introducing the Illusory Poisoning Attack against Neural Radiance Fields (IPA-NeRF). This attack involves embedding a hidden backdoor view into NeRF, allowing it to produce predetermined outputs, i.e. illusory, when presented with the specified backdoor view while maintaining normal performance with standard inputs. Our attack is specifically designed to deceive users or downstream models at a particular position while ensuring that any abnormalities in NeRF remain undetectable from other viewpoints. Experimental results demonstrate the effectiveness of our Illusory Poisoning Attack, successfully presenting the desired illusory on the specified viewpoint without impacting other views. Notably, we achieve this attack by introducing small perturbations solely to the training set. The code can be found at https://github.com/jiang-wenxiang/IPA-NeRF.
Photosequencing of Motion Blur using Short and Long Exposures
Dec 11, 2019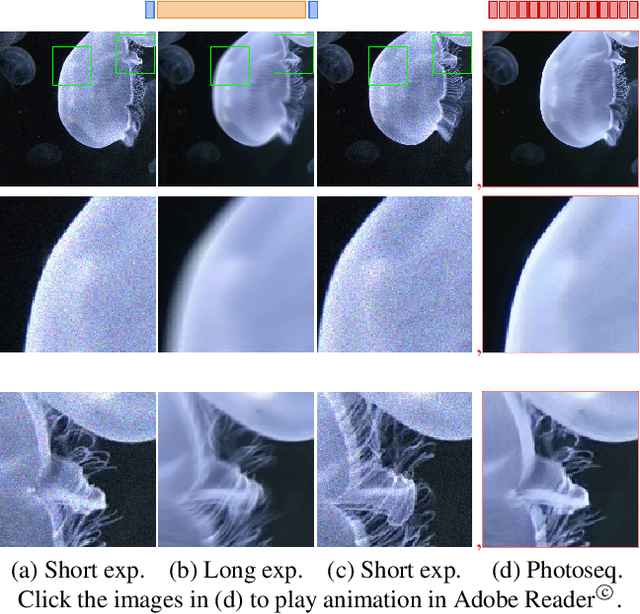

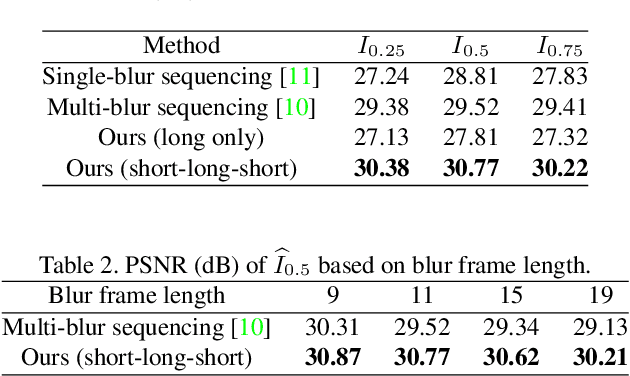
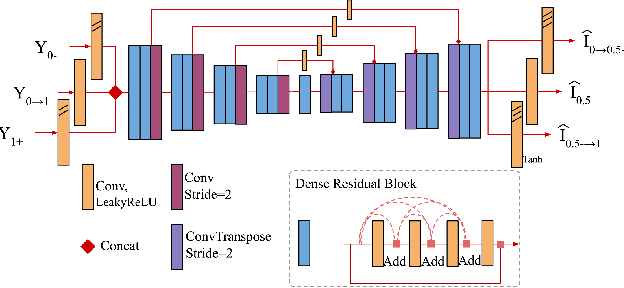
Abstract:Photosequencing aims to transform a motion blurred image to a sequence of sharp images. This problem is challenging due to the inherent ambiguities in temporal ordering as well as the recovery of lost spatial textures due to blur. Adopting a computational photography approach, we propose to capture two short exposure images, along with the original blurred long exposure image to aid in the aforementioned challenges. Post-capture, we recover the sharp photosequence using a novel blur decomposition strategy that recursively splits the long exposure image into smaller exposure intervals. We validate the approach by capturing a variety of scenes with interesting motions using machine vision cameras programmed to capture short and long exposure sequences. Our experimental results show that the proposed method resolves both fast and fine motions better than prior works.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge