Shimeng Huang
The Third Pillar of Causal Analysis? A Measurement Perspective on Causal Representations
May 23, 2025Abstract:Causal reasoning and discovery, two fundamental tasks of causal analysis, often face challenges in applications due to the complexity, noisiness, and high-dimensionality of real-world data. Despite recent progress in identifying latent causal structures using causal representation learning (CRL), what makes learned representations useful for causal downstream tasks and how to evaluate them are still not well understood. In this paper, we reinterpret CRL using a measurement model framework, where the learned representations are viewed as proxy measurements of the latent causal variables. Our approach clarifies the conditions under which learned representations support downstream causal reasoning and provides a principled basis for quantitatively assessing the quality of representations using a new Test-based Measurement EXclusivity (T-MEX) score. We validate T-MEX across diverse causal inference scenarios, including numerical simulations and real-world ecological video analysis, demonstrating that the proposed framework and corresponding score effectively assess the identification of learned representations and their usefulness for causal downstream tasks.
Supervised Learning and Model Analysis with Compositional Data
May 15, 2022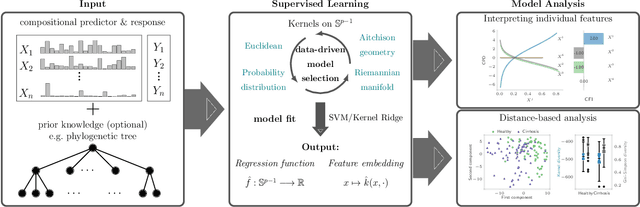
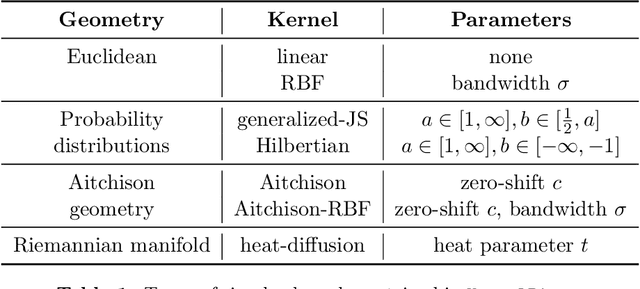
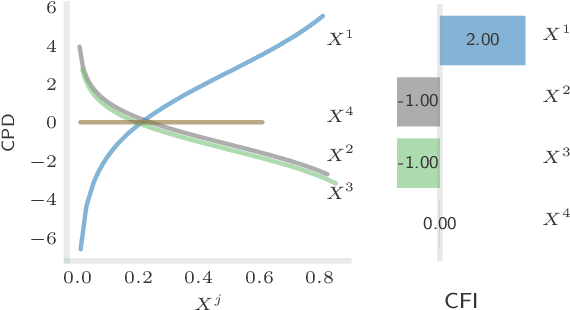
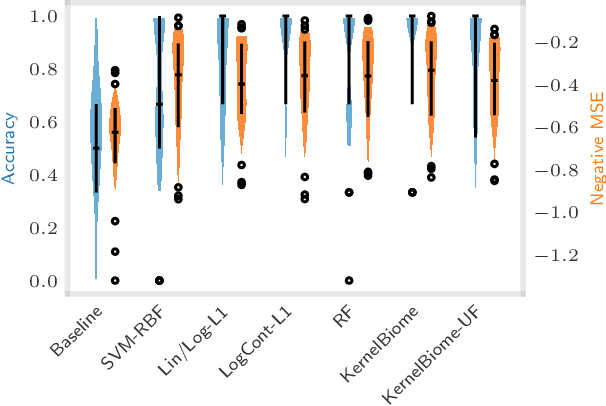
Abstract:The compositionality and sparsity of high-throughput sequencing data poses a challenge for regression and classification. However, in microbiome research in particular, conditional modeling is an essential tool to investigate relationships between phenotypes and the microbiome. Existing techniques are often inadequate: they either rely on extensions of the linear log-contrast model (which adjusts for compositionality, but is often unable to capture useful signals), or they are based on black-box machine learning methods (which may capture useful signals, but ignore compositionality in downstream analyses). We propose KernelBiome, a kernel-based nonparametric regression and classification framework for compositional data. It is tailored to sparse compositional data and is able to incorporate prior knowledge, such as phylogenetic structure. KernelBiome captures complex signals, including in the zero-structure, while automatically adapting model complexity. We demonstrate on par or improved predictive performance compared with state-of-the-art machine learning methods. Additionally, our framework provides two key advantages: (i) We propose two novel quantities to interpret contributions of individual components and prove that they consistently estimate average perturbation effects of the conditional mean, extending the interpretability of linear log-contrast models to nonparametric models. (ii) We show that the connection between kernels and distances aids interpretability and provides a data-driven embedding that can augment further analysis. Finally, we apply the KernelBiome framework to two public microbiome studies and illustrate the proposed model analysis. KernelBiome is available as an open-source Python package at https://github.com/shimenghuang/KernelBiome.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge