Sergio Serra
SYN-MAD 2022: Competition on Face Morphing Attack Detection Based on Privacy-aware Synthetic Training Data
Aug 15, 2022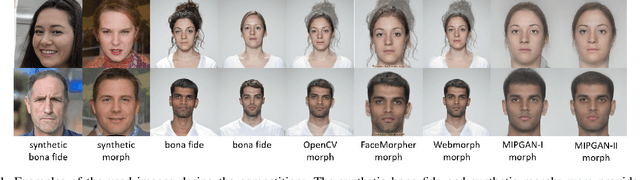
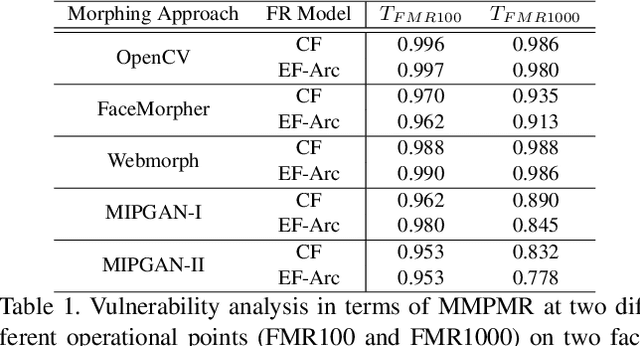
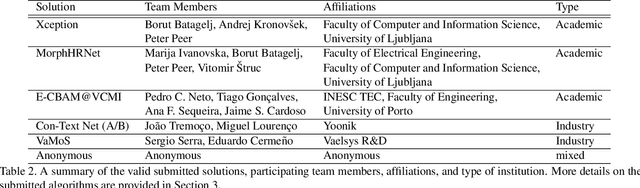
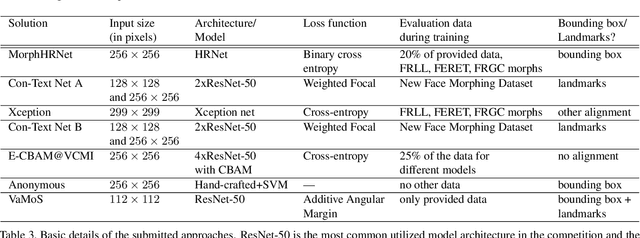
Abstract:This paper presents a summary of the Competition on Face Morphing Attack Detection Based on Privacy-aware Synthetic Training Data (SYN-MAD) held at the 2022 International Joint Conference on Biometrics (IJCB 2022). The competition attracted a total of 12 participating teams, both from academia and industry and present in 11 different countries. In the end, seven valid submissions were submitted by the participating teams and evaluated by the organizers. The competition was held to present and attract solutions that deal with detecting face morphing attacks while protecting people's privacy for ethical and legal reasons. To ensure this, the training data was limited to synthetic data provided by the organizers. The submitted solutions presented innovations that led to outperforming the considered baseline in many experimental settings. The evaluation benchmark is now available at: https://github.com/marcohuber/SYN-MAD-2022.
GAP Enhancing Semantic Interoperability of Genomic Datasets and Provenance Through Nanopublications
Nov 18, 2021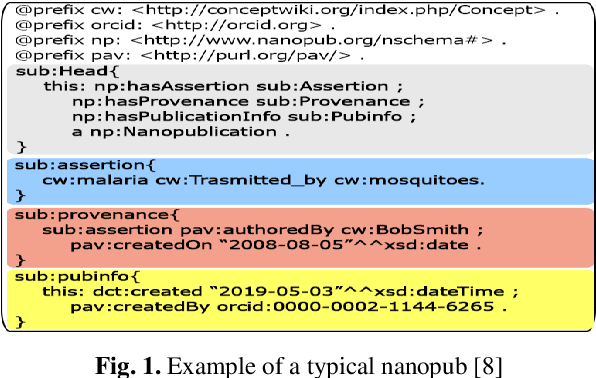
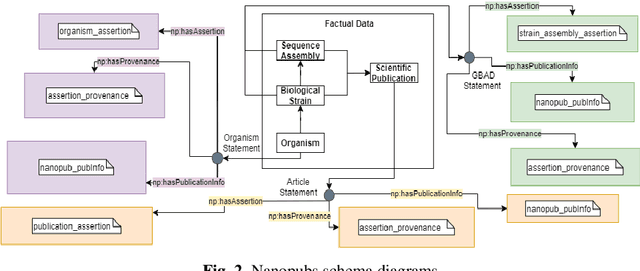

Abstract:While the publication of datasets in scientific repositories has become broadly recognised, the repositories tend to have increasing semantic-related problems. For instance, they present various data reuse obstacles for machine-actionable processes, especially in biological repositories, hampering the reproducibility of scientific experiments. An example of these shortcomings is the GenBank database. We propose GAP, an innovative data model to enhance the semantic data meaning to address these issues. The model focuses on converging related approaches like data provenance, semantic interoperability, FAIR principles, and nanopublications. Our experiments include a prototype to scrape genomic data and trace them to nanopublications as a proof of concept. For this, (meta)data are stored in a three-level nanopub data model. The first level is related to a target organism, specifying data in terms of biological taxonomy. The second level focuses on the biological strains of the target, the central part of our contribution. The strains express information related to deciphered (meta)data of the genetic variations of the genomic material. The third level stores related scientific papers (meta)data. We expect it will offer higher data storage flexibility and more extensive interoperability with other data sources by incorporating and adopting associated approaches to store genomic data in the proposed model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge