Sergi Valverde
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021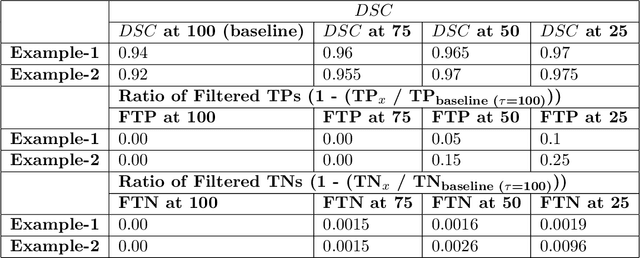
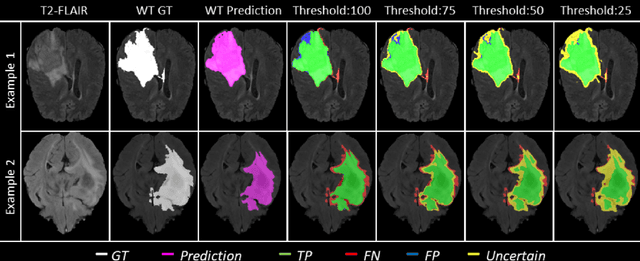
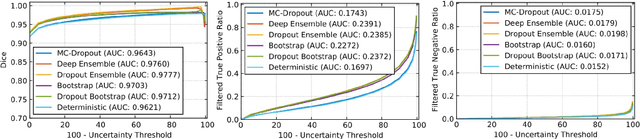
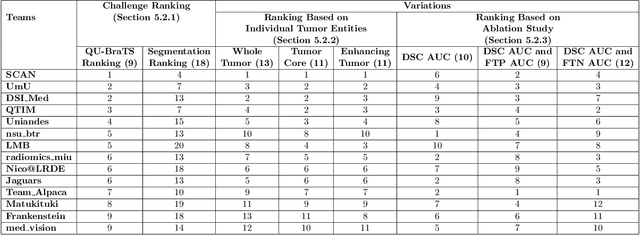
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019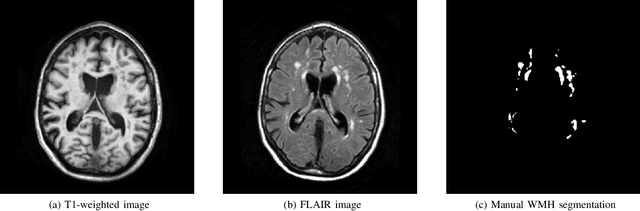
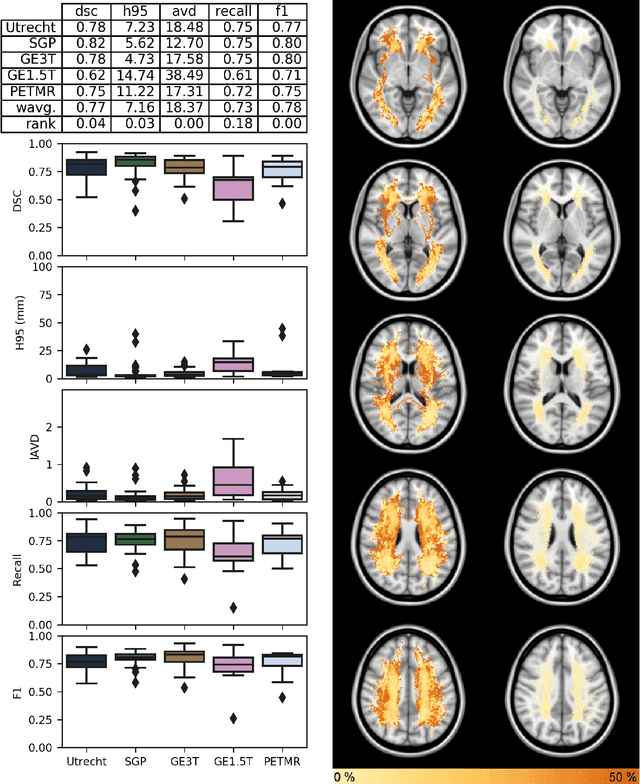
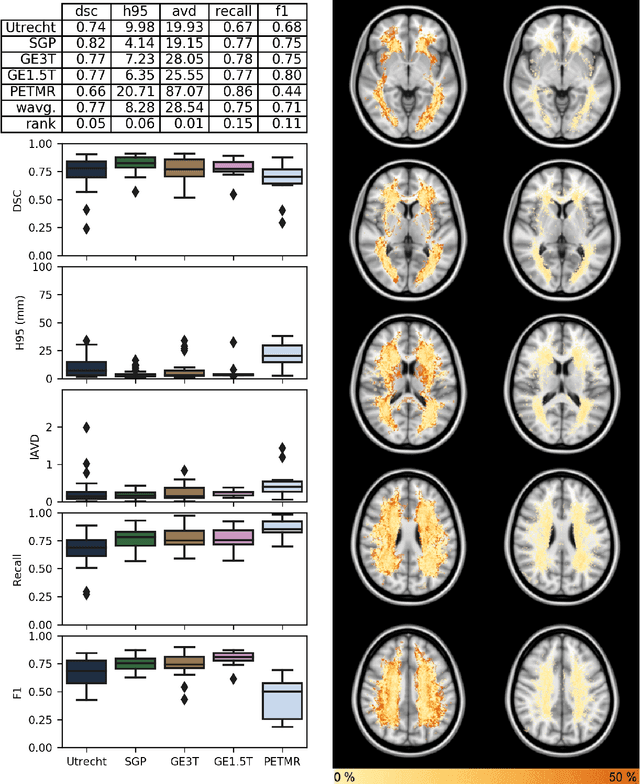
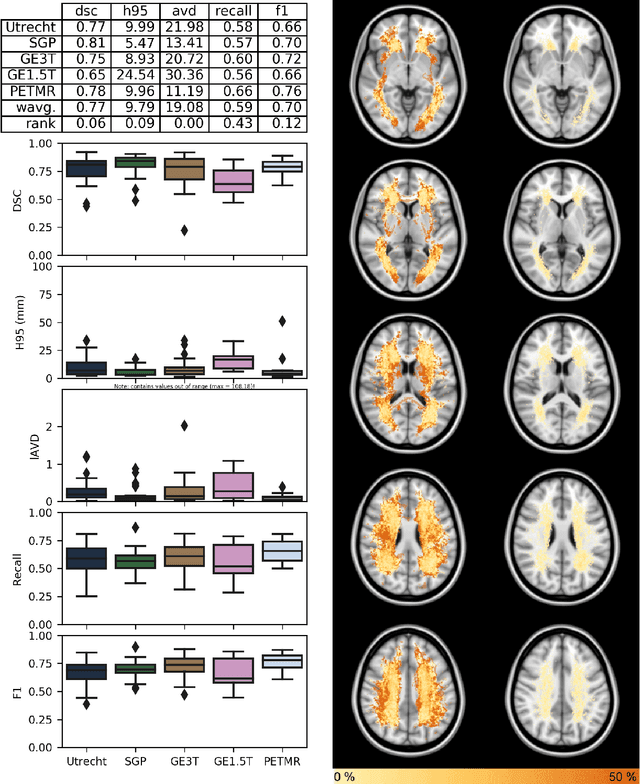
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
Multiple Sclerosis Lesion Synthesis in MRI using an encoder-decoder U-NET
Jan 17, 2019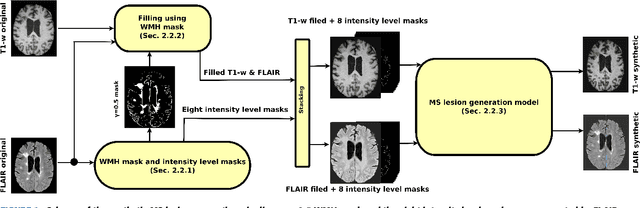

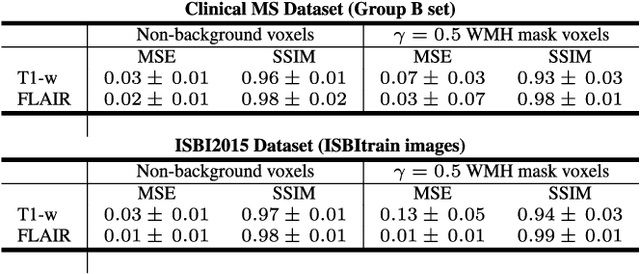
Abstract:In this paper, we propose generating synthetic multiple sclerosis (MS) lesions on MRI images with the final aim to improve the performance of supervised machine learning algorithms, therefore avoiding the problem of the lack of available ground truth. We propose a two-input two-output fully convolutional neural network model for MS lesion synthesis in MRI images. The lesion information is encoded as discrete binary intensity level masks passed to the model and stacked with the input images. The model is trained end-to-end without the need for manually annotating the lesions in the training set. We then perform the generation of synthetic lesions on healthy images via registration of patient images, which are subsequently used for data augmentation to increase the performance for supervised MS lesion detection algorithms. Our pipeline is evaluated on MS patient data from an in-house clinical dataset and the public ISBI2015 challenge dataset. The evaluation is based on measuring the similarities between the real and the synthetic images as well as in terms of lesion detection performance by segmenting both the original and synthetic images individually using a state-of-the-art segmentation framework. We also demonstrate the usage of synthetic MS lesions generated on healthy images as data augmentation. We analyze a scenario of limited training data (one-image training) to demonstrate the effect of the data augmentation on both datasets. Our results significantly show the effectiveness of the usage of synthetic MS lesion images. For the ISBI2015 challenge, our one-image model trained using only a single image plus the synthetic data augmentation strategy showed a performance similar to that of other CNN methods that were fully trained using the entire training set, yielding a comparable human expert rater performance
SUNet: a deep learning architecture for acute stroke lesion segmentation and outcome prediction in multimodal MRI
Oct 31, 2018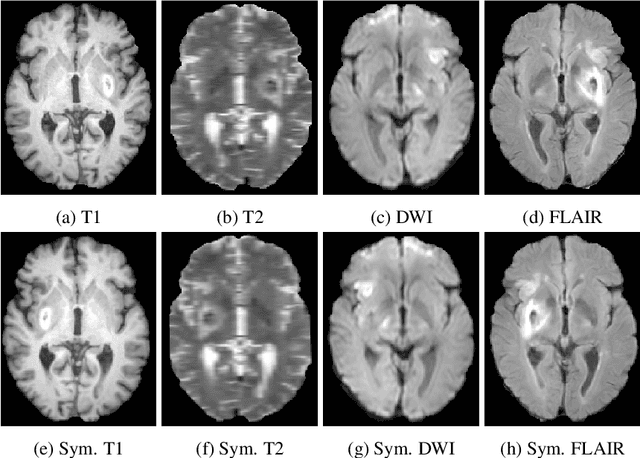
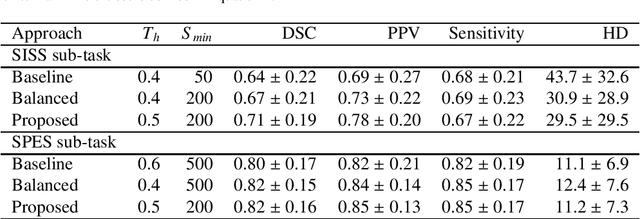
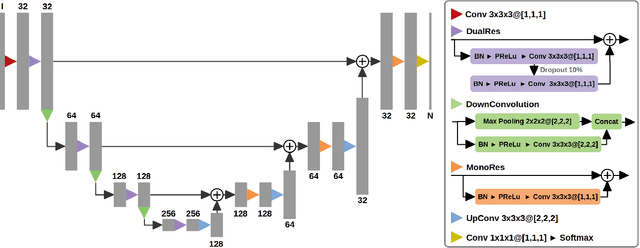
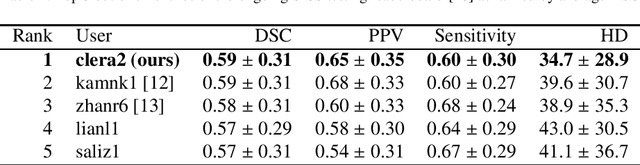
Abstract:Acute stroke lesion segmentation and prediction tasks are of great clinical interest as they can help doctors make better informed time-critical treatment decisions. Automatic segmentation of these lesions is a complex task due to their heterogeneous appearance, dynamic evolution and inter-patient differences. Typically, acute stroke lesion tasks are approached with methods developed for chronic stroke or other brain lesions. However, the pathophysiology and anatomy of acute stroke establishes an inherently different problem that needs special consideration. In this work, we propose a novel deep learning architecture specially designed for acute stroke tasks that involve approximating complex non-linear functions with reduced data. Within our strategy, class imbalance is tackled using a hybrid strategy based on state-of-the-art train sampling strategies designed for other brain lesion related tasks, which is more suited to the anatomy and pathophysiology of acute stroke lesions. The proposed method is evaluated on three unrelated public international challenge datasets (ISLES) without any dataset specific hyper-parameter tuning. These involve the tasks of sub-acute stroke lesion segmentation, acute stroke penumbra estimation and chronic extent prediction from acute MR images. The performance of the proposed architecture is analysed both against similar deep learning architectures from chronic stroke and related biomedical tasks and also by submitting the segmented test images for blind online evaluation on each of the challenges. When compared with the rest of submitted strategies, our method achieves top-rank performance among the best submitted entries in all the three challenges, showing its capability to deal with different unrelated tasks without hyper-parameter tuning. In order to promote the reproducibility of our results, a public version of the proposed method has been released.
Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review
Jun 11, 2018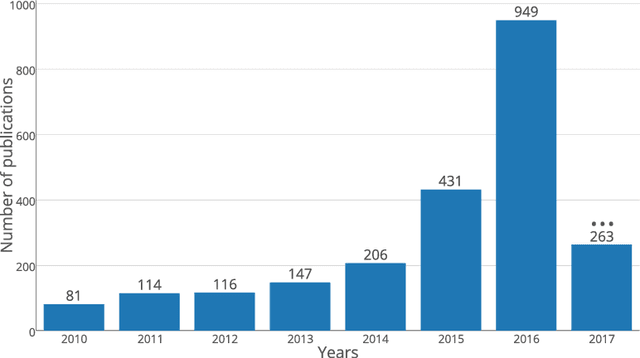
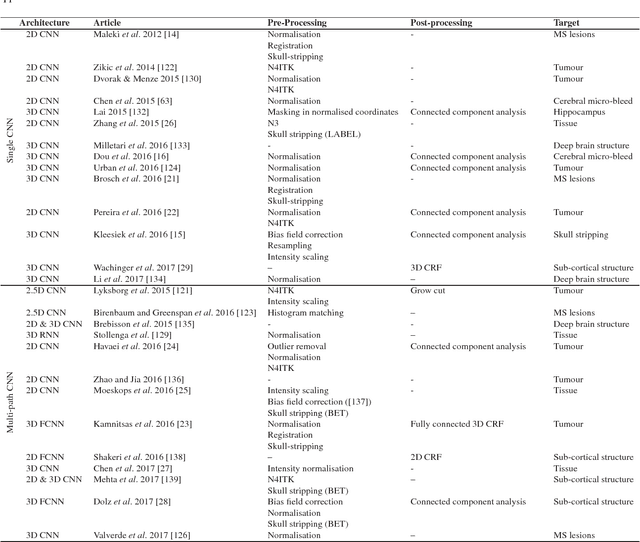
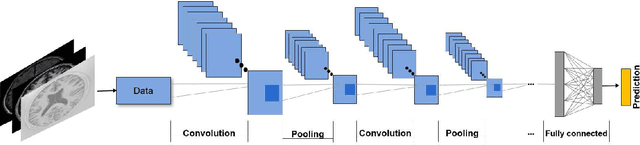
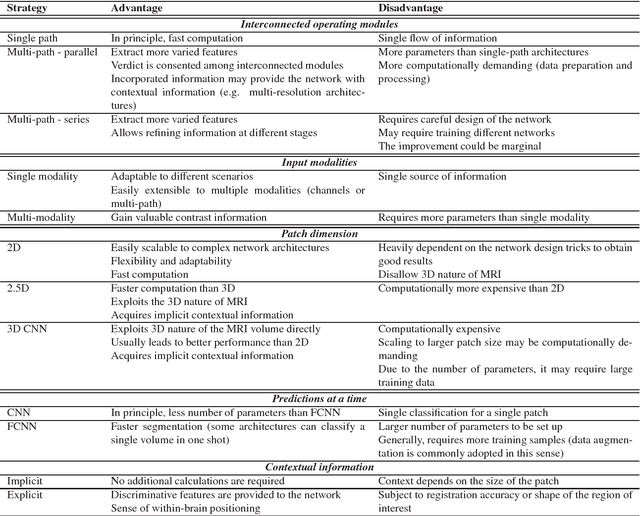
Abstract:In recent years, deep convolutional neural networks (CNNs) have shown record-shattering performance in a variety of computer vision problems, such as visual object recognition, detection and segmentation. These methods have also been utilised in medical image analysis domain for lesion segmentation, anatomical segmentation and classification. We present an extensive literature review of CNN techniques applied in brain magnetic resonance imaging (MRI) analysis, focusing on the architectures, pre-processing, data-preparation and post-processing strategies available in these works. The aim of this study is three-fold. Our primary goal is to report how different CNN architectures have evolved, discuss state-of-the-art strategies, condense their results obtained using public datasets and examine their pros and cons. Second, this paper is intended to be a detailed reference of the research activity in deep CNN for brain MRI analysis. Finally, we present a perspective on the future of CNNs in which we hint some of the research directions in subsequent years.
One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks
May 31, 2018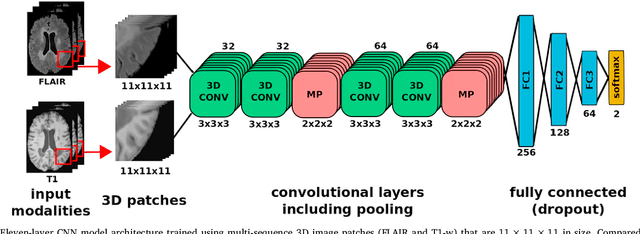
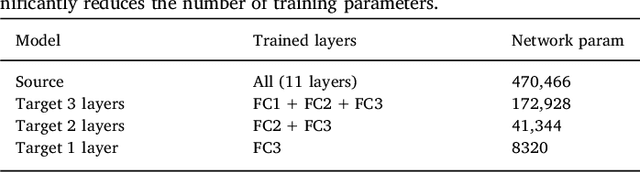
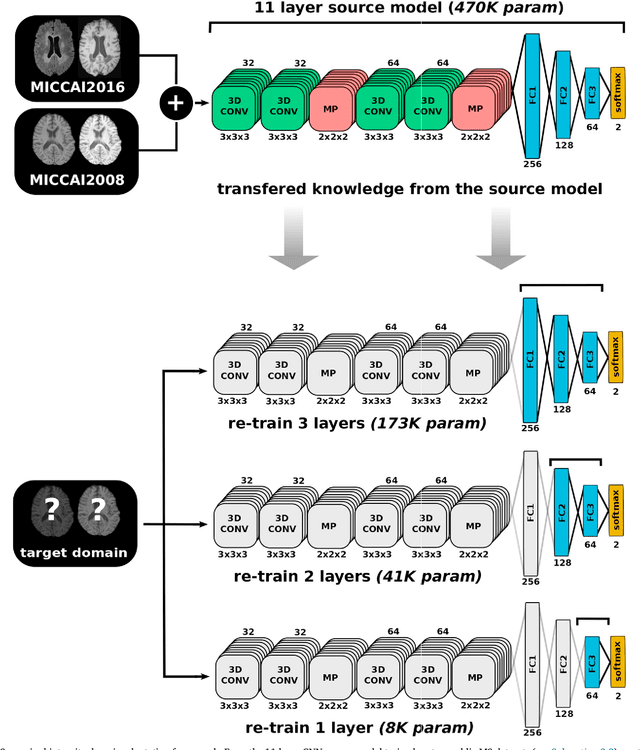
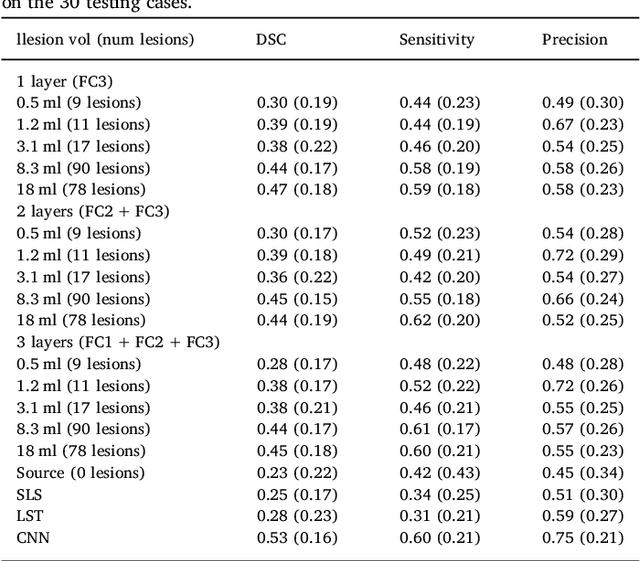
Abstract:In recent years, several convolutional neural network (CNN) methods have been proposed for the automated white matter lesion segmentation of multiple sclerosis (MS) patient images, due to their superior performance compared with those of other state-of-the-art methods. However, the accuracies of CNN methods tend to decrease significantly when evaluated on different image domains compared with those used for training, which demonstrates the lack of adaptability of CNNs to unseen imaging data. In this study, we analyzed the effect of intensity domain adaptation on our recently proposed CNN-based MS lesion segmentation method. Given a source model trained on two public MS datasets, we investigated the transferability of the CNN model when applied to other MRI scanners and protocols, evaluating the minimum number of annotated images needed from the new domain and the minimum number of layers needed to re-train to obtain comparable accuracy. Our analysis comprised MS patient data from both a clinical center and the public ISBI2015 challenge database, which permitted us to compare the domain adaptation capability of our model to that of other state-of-the-art methods. For the ISBI2015 challenge, our one-shot domain adaptation model trained using only a single image showed a performance similar to that of other CNN methods that were fully trained using the entire available training set, yielding a comparable human expert rater performance. We believe that our experiments will encourage the MS community to incorporate its use in different clinical settings with reduced amounts of annotated data. This approach could be meaningful not only in terms of the accuracy in delineating MS lesions but also in the related reductions in time and economic costs derived from manual lesion labeling.
Quantitative analysis of patch-based fully convolutional neural networks for tissue segmentation on brain magnetic resonance imaging
Feb 19, 2018
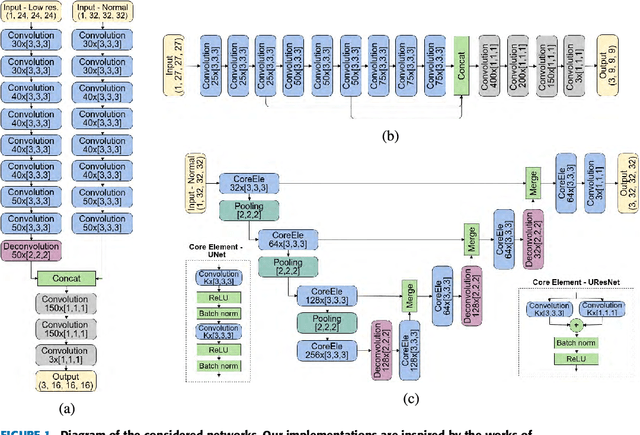
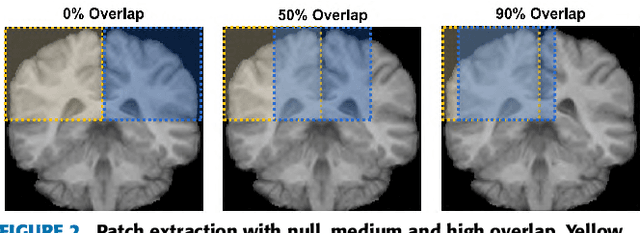
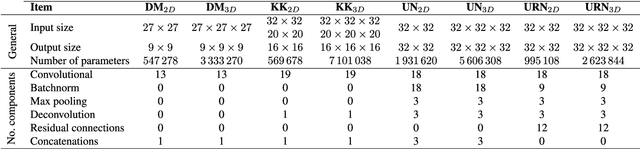
Abstract:Accurate brain tissue segmentation in Magnetic Resonance Imaging (MRI) has attracted the attention of medical doctors and researchers since variations in tissue volume help in diagnosing and monitoring neurological diseases. Several proposals have been designed throughout the years comprising conventional machine learning strategies as well as convolutional neural networks (CNN) approaches. In particular, in this paper, we analyse a sub-group of deep learning methods producing dense predictions. This branch, referred in the literature as Fully CNN (FCNN), is of interest as these architectures can process an input volume in less time than CNNs and local spatial dependencies may be encoded since several voxels are classified at once. Our study focuses on understanding architectural strengths and weaknesses of literature-like approaches. Hence, we implement eight FCNN architectures inspired by robust state-of-the-art methods on brain segmentation related tasks. We evaluate them using the IBSR18, MICCAI2012 and iSeg2017 datasets as they contain infant and adult data and exhibit varied voxel spacing, image quality, number of scans and available imaging modalities. The discussion is driven in three directions: comparison between 2D and 3D approaches, the importance of multiple modalities and overlapping as a sampling strategy for training and testing models. To encourage other researchers to explore the evaluation framework, a public version is accessible to download from our research website.
Automated sub-cortical brain structure segmentation combining spatial and deep convolutional features
Sep 26, 2017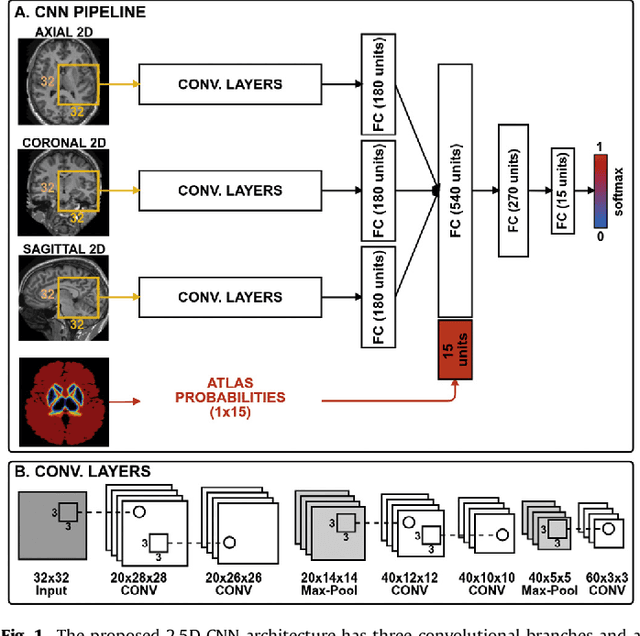
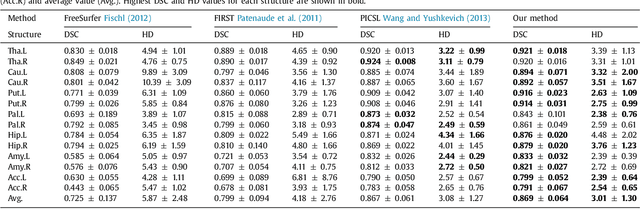
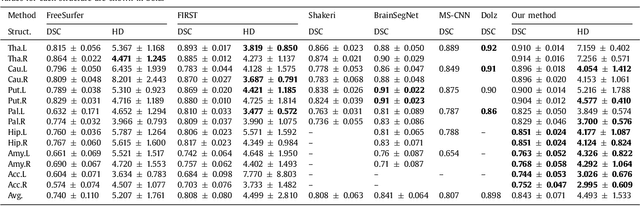
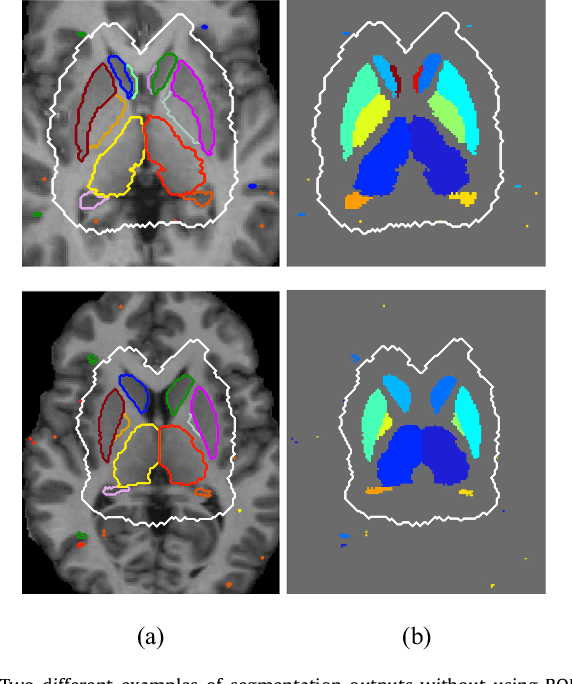
Abstract:Sub-cortical brain structure segmentation in Magnetic Resonance Images (MRI) has attracted the interest of the research community for a long time because morphological changes in these structures are related to different neurodegenerative disorders. However, manual segmentation of these structures can be tedious and prone to variability, highlighting the need for robust automated segmentation methods. In this paper, we present a novel convolutional neural network based approach for accurate segmentation of the sub-cortical brain structures that combines both convolutional and prior spatial features for improving the segmentation accuracy. In order to increase the accuracy of the automated segmentation, we propose to train the network using a restricted sample selection to force the network to learn the most difficult parts of the structures. We evaluate the accuracy of the proposed method on the public MICCAI 2012 challenge and IBSR 18 datasets, comparing it with different available state-of-the-art methods and other recently proposed deep learning approaches. On the MICCAI 2012 dataset, our method shows an excellent performance comparable to the best challenge participant strategy, while performing significantly better than state-of-the-art techniques such as FreeSurfer and FIRST. On the IBSR 18 dataset, our method also exhibits a significant increase in the performance with respect to not only FreeSurfer and FIRST, but also comparable or better results than other recent deep learning approaches. Moreover, our experiments show that both the addition of the spatial priors and the restricted sampling strategy have a significant effect on the accuracy of the proposed method. In order to encourage the reproducibility and the use of the proposed method, a public version of our approach is available to download for the neuroimaging community.
Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach
Feb 16, 2017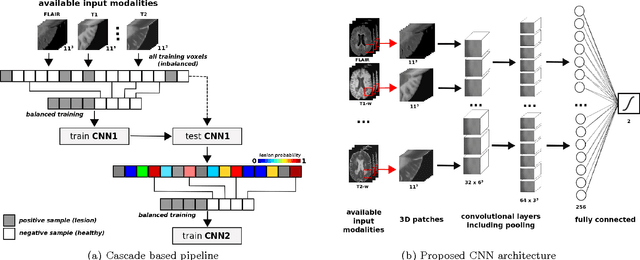
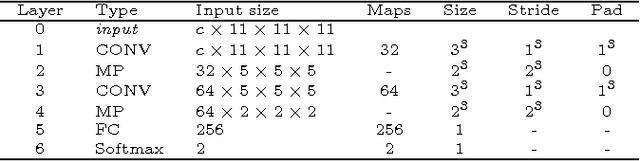
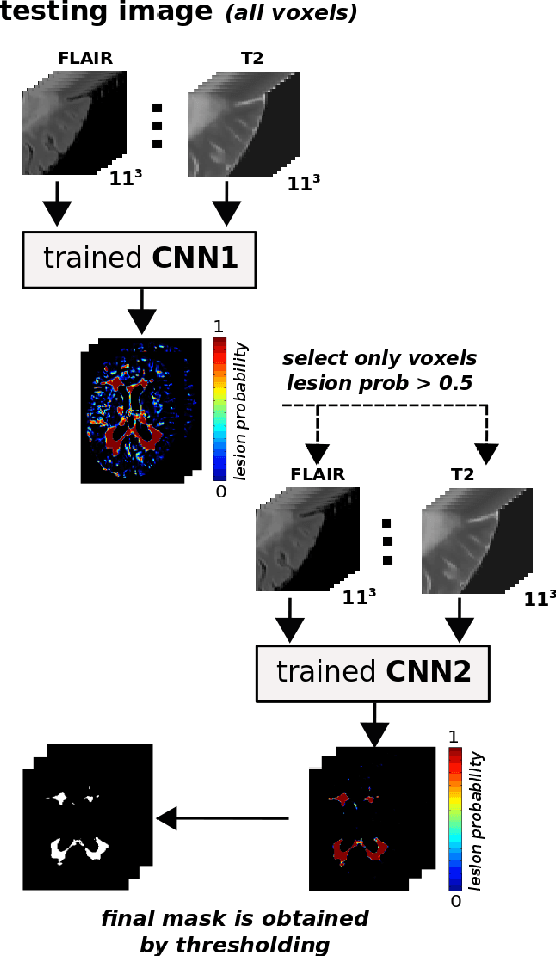
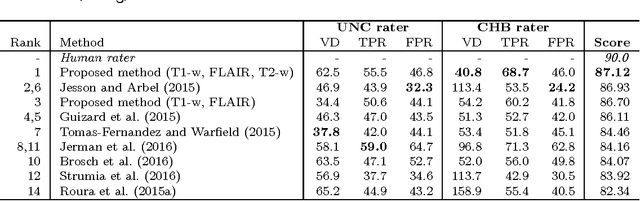
Abstract:In this paper, we present a novel automated method for White Matter (WM) lesion segmentation of Multiple Sclerosis (MS) patient images. Our approach is based on a cascade of two 3D patch-wise convolutional neural networks (CNN). The first network is trained to be more sensitive revealing possible candidate lesion voxels while the second network is trained to reduce the number of misclassified voxels coming from the first network. This cascaded CNN architecture tends to learn well from small sets of training data, which can be very interesting in practice, given the difficulty to obtain manual label annotations and the large amount of available unlabeled Magnetic Resonance Imaging (MRI) data. We evaluate the accuracy of the proposed method on the public MS lesion segmentation challenge MICCAI2008 dataset, comparing it with respect to other state-of-the-art MS lesion segmentation tools. Furthermore, the proposed method is also evaluated on two private MS clinical datasets, where the performance of our method is also compared with different recent public available state-of-the-art MS lesion segmentation methods. At the time of writing this paper, our method is the best ranked approach on the MICCAI2008 challenge, outperforming the rest of 60 participant methods when using all the available input modalities (T1-w, T2-w and FLAIR), while still in the top-rank (3rd position) when using only T1-w and FLAIR modalities. On clinical MS data, our approach exhibits a significant increase in the accuracy segmenting of WM lesions when compared with the rest of evaluated methods, highly correlating ($r \ge 0.97$) also with the expected lesion volume.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge