Satish E. Viswanath
Information-driven Fusion of Pathology Foundation Models for Enhanced Disease Characterization
Dec 11, 2025Abstract:Foundation models (FMs) have demonstrated strong performance across diverse pathology tasks. While there are similarities in the pre-training objectives of FMs, there is still limited understanding of their complementarity, redundancy in embedding spaces, or biological interpretation of features. In this study, we propose an information-driven, intelligent fusion strategy for integrating multiple pathology FMs into a unified representation and systematically evaluate its performance for cancer grading and staging across three distinct diseases. Diagnostic H&E whole-slide images from kidney (519 slides), prostate (490 slides), and rectal (200 slides) cancers were dichotomized into low versus high grade or stage. Both tile-level FMs (Conch v1.5, MUSK, Virchow2, H-Optimus1, Prov-Gigapath) and slide-level FMs (TITAN, CHIEF, MADELEINE) were considered to train downstream classifiers. We then evaluated three FM fusion schemes at both tile and slide levels: majority-vote ensembling, naive feature concatenation, and intelligent fusion based on correlation-guided pruning of redundant features. Under patient-stratified cross-validation with hold-out testing, intelligent fusion of tile-level embeddings yielded consistent gains in classification performance across all three cancers compared with the best single FMs and naive fusion. Global similarity metrics revealed substantial alignment of FM embedding spaces, contrasted by lower local neighborhood agreement, indicating complementary fine-grained information across FMs. Attention maps showed that intelligent fusion yielded concentrated attention on tumor regions while reducing spurious focus on benign regions. Our findings suggest that intelligent, correlation-guided fusion of pathology FMs can yield compact, task-tailored representations that enhance both predictive performance and interpretability in downstream computational pathology tasks.
CohortFinder: an open-source tool for data-driven partitioning of biomedical image cohorts to yield robust machine learning models
Jul 17, 2023

Abstract:Batch effects (BEs) refer to systematic technical differences in data collection unrelated to biological variations whose noise is shown to negatively impact machine learning (ML) model generalizability. Here we release CohortFinder, an open-source tool aimed at mitigating BEs via data-driven cohort partitioning. We demonstrate CohortFinder improves ML model performance in downstream medical image processing tasks. CohortFinder is freely available for download at cohortfinder.com.
MRQy: An Open-Source Tool for Quality Control of MR Imaging Data
Apr 13, 2020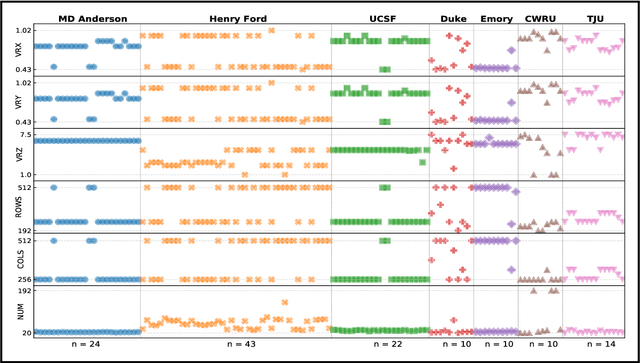
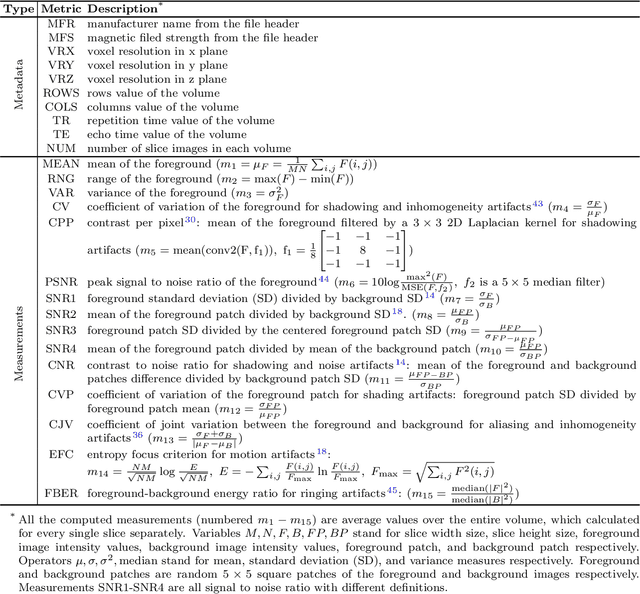
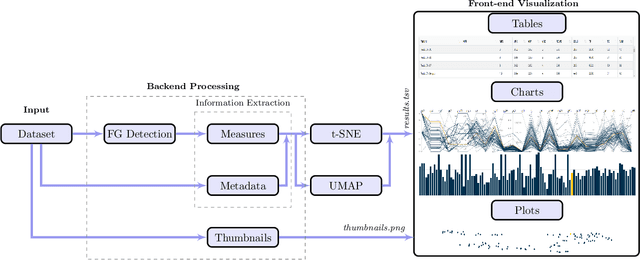
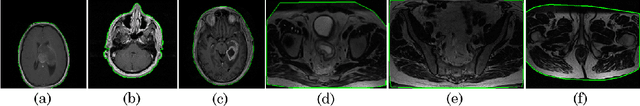
Abstract:Even as public data repositories such as The Cancer Imaging Archive (TCIA) have enabled development of new radiomics and machine learning schemes, a key concern remains the generalizability of these methods to unseen datasets. For MRI datasets, model performance could be impacted by (a) site- or scanner-specific variations in image resolution, field-of-view, or image contrast, or (b) presence of imaging artifacts such as noise, motion, inhomogeneity, ringing, or aliasing; which can adversely affect relative image quality between data cohorts. This indicates a need for a quantitative tool to quickly determine relative differences in MRI volumes both within and between large data cohorts. We present MRQy, a new open-source quality control tool to (a) interrogate MRI cohorts for site- or equipment-based differences, and (b) quantify the impact of MRI artifacts on relative image quality; to help determine how to correct for these variations prior to model development. MRQy extracts a series of quality measures (e.g. noise ratios, variation metrics, entropy and energy criteria) and MR image metadata (e.g. voxel resolution, image dimensions) for subsequent interrogation via a specialized HTML5 based front-end designed for real-time filtering and trend visualization. MRQy is designed to be a standalone, unsupervised tool that can be efficiently run on a standard desktop computer. It has been made freely accessible at http://github.com/ccipd/MRQy for wider community use and feedback. MRQy was used to evaluate (a) n=133 brain MRIs from TCIA (7 sites), and (b) n=104 rectal MRIs (3 local sites). MRQy measures revealed significant site-specific variations in both cohorts, indicating potential batch effects. Marked differences in specific MRQy measures were also able to identify MRI datasets that needed to be corrected for common MR imaging artifacts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge