Santiago Romero-Brufau
BRIDGE: Benchmarking Large Language Models for Understanding Real-world Clinical Practice Text
May 01, 2025



Abstract:Large language models (LLMs) hold great promise for medical applications and are evolving rapidly, with new models being released at an accelerated pace. However, current evaluations of LLMs in clinical contexts remain limited. Most existing benchmarks rely on medical exam-style questions or PubMed-derived text, failing to capture the complexity of real-world electronic health record (EHR) data. Others focus narrowly on specific application scenarios, limiting their generalizability across broader clinical use. To address this gap, we present BRIDGE, a comprehensive multilingual benchmark comprising 87 tasks sourced from real-world clinical data sources across nine languages. We systematically evaluated 52 state-of-the-art LLMs (including DeepSeek-R1, GPT-4o, Gemini, and Llama 4) under various inference strategies. With a total of 13,572 experiments, our results reveal substantial performance variation across model sizes, languages, natural language processing tasks, and clinical specialties. Notably, we demonstrate that open-source LLMs can achieve performance comparable to proprietary models, while medically fine-tuned LLMs based on older architectures often underperform versus updated general-purpose models. The BRIDGE and its corresponding leaderboard serve as a foundational resource and a unique reference for the development and evaluation of new LLMs in real-world clinical text understanding. The BRIDGE leaderboard: https://huggingface.co/spaces/YLab-Open/BRIDGE-Medical-Leaderboard
DeepABM: Scalable, efficient and differentiable agent-based simulations via graph neural networks
Oct 09, 2021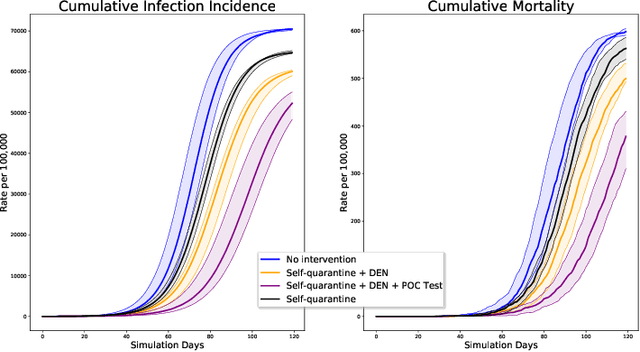

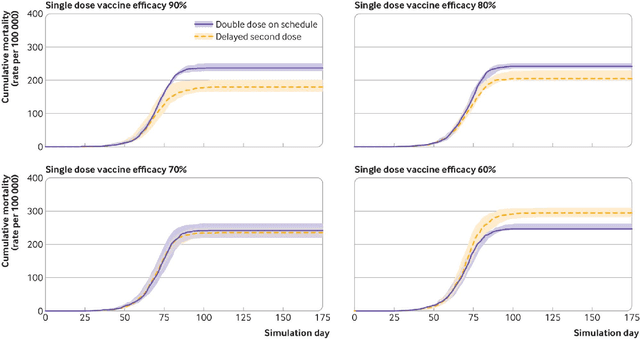
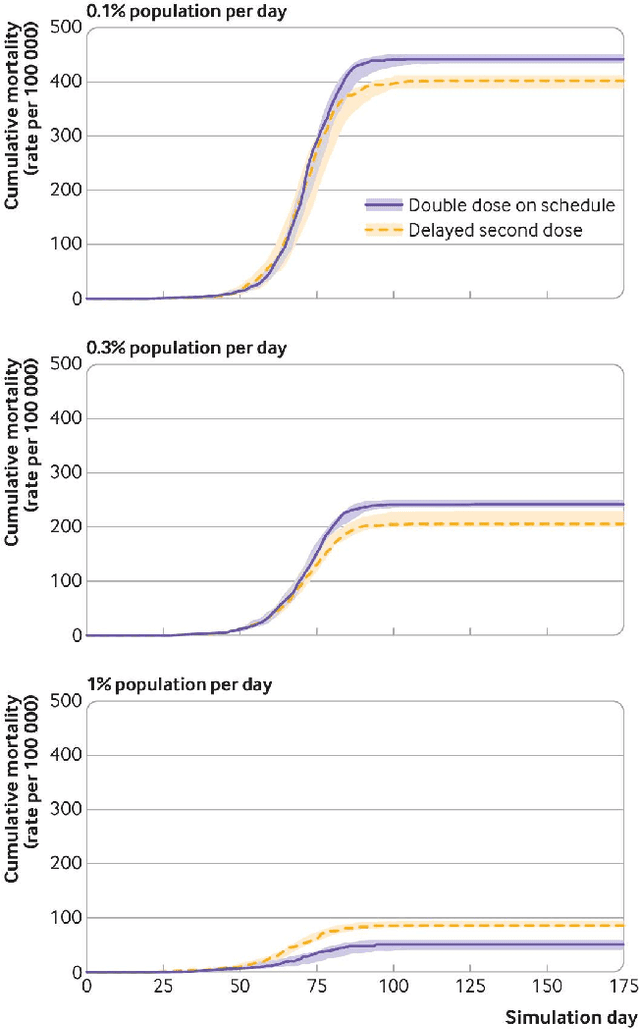
Abstract:We introduce DeepABM, a framework for agent-based modeling that leverages geometric message passing of graph neural networks for simulating action and interactions over large agent populations. Using DeepABM allows scaling simulations to large agent populations in real-time and running them efficiently on GPU architectures. To demonstrate the effectiveness of DeepABM, we build DeepABM-COVID simulator to provide support for various non-pharmaceutical interventions (quarantine, exposure notification, vaccination, testing) for the COVID-19 pandemic, and can scale to populations of representative size in real-time on a GPU. Specifically, DeepABM-COVID can model 200 million interactions (over 100,000 agents across 180 time-steps) in 90 seconds, and is made available online to help researchers with modeling and analysis of various interventions. We explain various components of the framework and discuss results from one research study to evaluate the impact of delaying the second dose of the COVID-19 vaccine in collaboration with clinical and public health experts. While we simulate COVID-19 spread, the ideas introduced in the paper are generic and can be easily extend to other forms of agent-based simulations. Furthermore, while beyond scope of this document, DeepABM enables inverse agent-based simulations which can be used to learn physical parameters in the (micro) simulations using gradient-based optimization with large-scale real-world (macro) data. We are optimistic that the current work can have interesting implications for bringing ABM and AI communities closer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge