Samaneh Samiei
Boosting multi-demographic federated learning for chest x-ray analysis using general-purpose self-supervised representations
Apr 11, 2025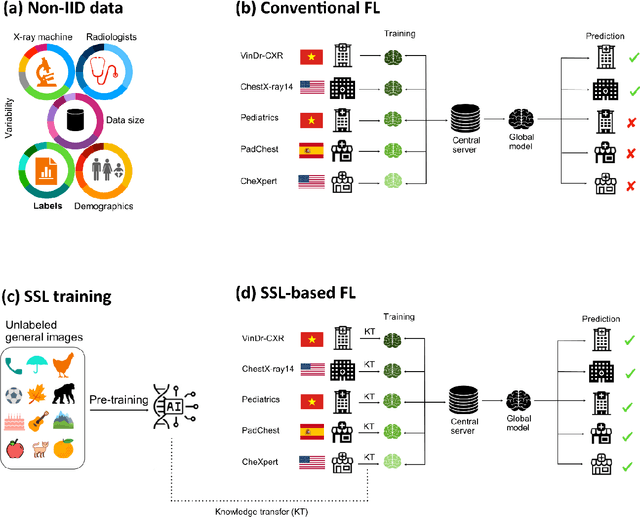



Abstract:Reliable artificial intelligence (AI) models for medical image analysis often depend on large and diverse labeled datasets. Federated learning (FL) offers a decentralized and privacy-preserving approach to training but struggles in highly non-independent and identically distributed (non-IID) settings, where institutions with more representative data may experience degraded performance. Moreover, existing large-scale FL studies have been limited to adult datasets, neglecting the unique challenges posed by pediatric data, which introduces additional non-IID variability. To address these limitations, we analyzed n=398,523 adult chest radiographs from diverse institutions across multiple countries and n=9,125 pediatric images, leveraging transfer learning from general-purpose self-supervised image representations to classify pneumonia and cases with no abnormality. Using state-of-the-art vision transformers, we found that FL improved performance only for smaller adult datasets (P<0.001) but degraded performance for larger datasets (P<0.064) and pediatric cases (P=0.242). However, equipping FL with self-supervised weights significantly enhanced outcomes across pediatric cases (P=0.031) and most adult datasets (P<0.008), except the largest dataset (P=0.052). These findings underscore the potential of easily deployable general-purpose self-supervised image representations to address non-IID challenges in clinical FL applications and highlight their promise for enhancing patient outcomes and advancing pediatric healthcare, where data scarcity and variability remain persistent obstacles.
A complex network approach to time series analysis with application in diagnosis of neuromuscular disorders
Aug 16, 2021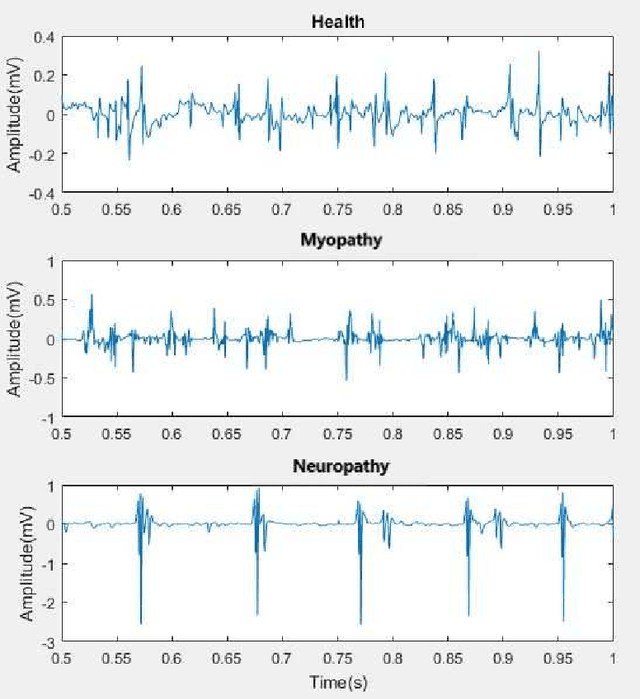
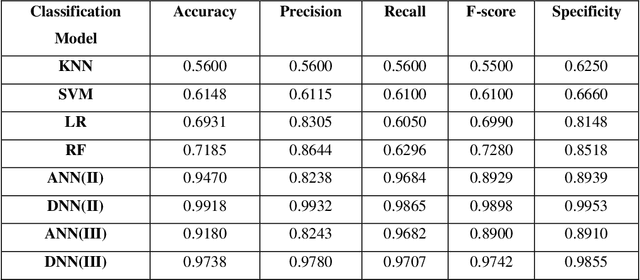
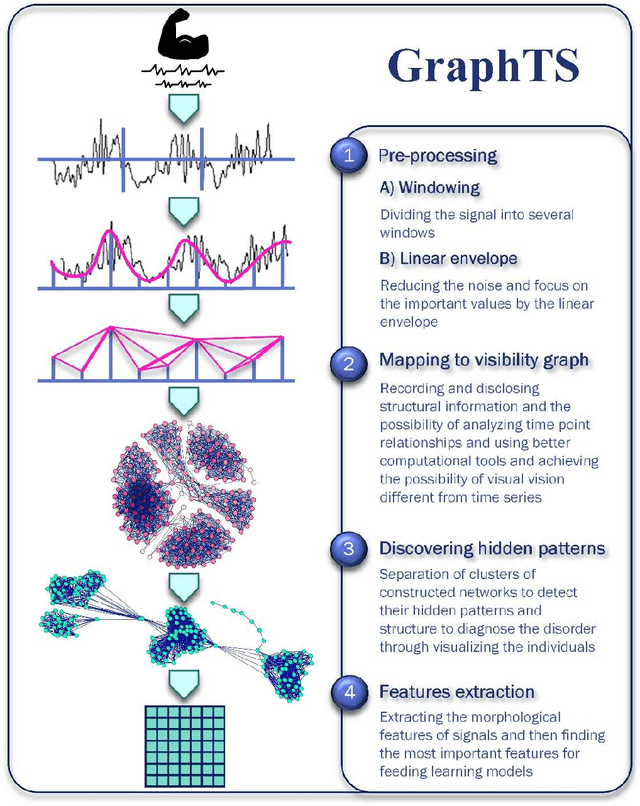
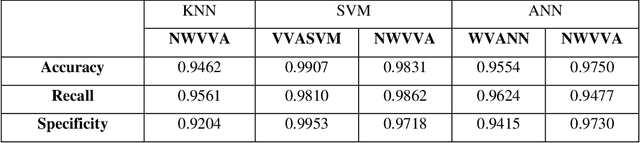
Abstract:Electromyography (EMG) refers to a biomedical signal indicating neuromuscular activity and muscle morphology. Experts accurately diagnose neuromuscular disorders using this time series. Modern data analysis techniques have recently led to introducing novel approaches for mapping time series data to graphs and complex networks with applications in diverse fields, including medicine. The resulting networks develop a completely different visual acuity that can be used to complement physician findings of time series. This can lead to a more enriched analysis, reduced error, more accurate diagnosis of the disease, and increased accuracy and speed of the treatment process. The mapping process may cause the loss of essential data from the time series and not retain all the time series features. As a result, achieving an approach that can provide a good representation of the time series while maintaining essential features is crucial. This paper proposes a new approach to network development named GraphTS to overcome the limited accuracy of existing methods through EMG time series using the visibility graph method. For this purpose, EMG signals are pre-processed and mapped to a complex network by a standard visibility graph algorithm. The resulting networks can differentiate between healthy and patient samples. In the next step, the properties of the developed networks are given in the form of a feature matrix as input to classifiers after extracting optimal features. Performance evaluation of the proposed approach with deep neural network shows 99.30% accuracy for training data and 99.18% for test data. Therefore, in addition to enriched network representation and covering the features of time series for healthy, myopathy, and neuropathy EMG, the proposed technique improves accuracy, precision, recall, and F-score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge