Salvatore Romano
Beyond MMD: Evaluating Graph Generative Models with Geometric Deep Learning
Dec 16, 2025



Abstract:Graph generation is a crucial task in many fields, including network science and bioinformatics, as it enables the creation of synthetic graphs that mimic the properties of real-world networks for various applications. Graph Generative Models (GGMs) have emerged as a promising solution to this problem, leveraging deep learning techniques to learn the underlying distribution of real-world graphs and generate new samples that closely resemble them. Examples include approaches based on Variational Auto-Encoders, Recurrent Neural Networks, and more recently, diffusion-based models. However, the main limitation often lies in the evaluation process, which typically relies on Maximum Mean Discrepancy (MMD) as a metric to assess the distribution of graph properties in the generated ensemble. This paper introduces a novel methodology for evaluating GGMs that overcomes the limitations of MMD, which we call RGM (Representation-aware Graph-generation Model evaluation). As a practical demonstration of our methodology, we present a comprehensive evaluation of two state-of-the-art Graph Generative Models: Graph Recurrent Attention Networks (GRAN) and Efficient and Degree-guided graph GEnerative model (EDGE). We investigate their performance in generating realistic graphs and compare them using a Geometric Deep Learning model trained on a custom dataset of synthetic and real-world graphs, specifically designed for graph classification tasks. Our findings reveal that while both models can generate graphs with certain topological properties, they exhibit significant limitations in preserving the structural characteristics that distinguish different graph domains. We also highlight the inadequacy of Maximum Mean Discrepancy as an evaluation metric for GGMs and suggest alternative approaches for future research.
Conditioning Normalizing Flows for Rare Event Sampling
Jul 29, 2022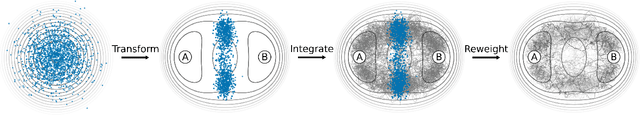
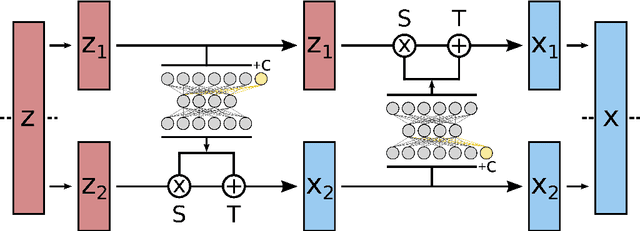
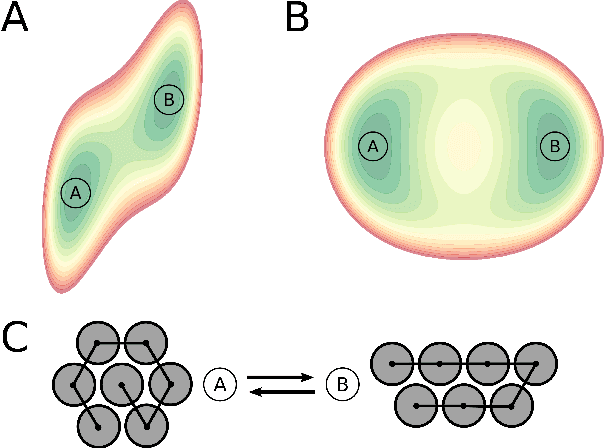
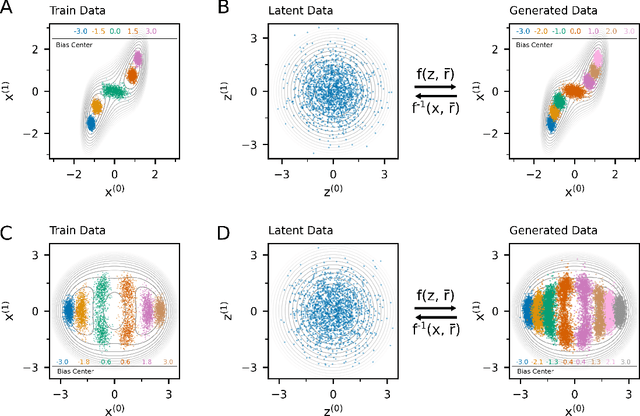
Abstract:Understanding the dynamics of complex molecular processes is often linked to the study of infrequent transitions between long-lived stable states. The standard approach to the sampling of such rare events is to generate an ensemble of transition paths using a random walk in trajectory space. This, however, comes with the drawback of strong correlation between subsequently visited paths and with an intrinsic difficulty in parallelizing the sampling process. We propose a transition path sampling scheme based on neural-network generated configurations. These are obtained employing normalizing flows, a neural network class able to generate decorrelated samples from a given distribution. With this approach, not only are correlations between visited paths removed, but the sampling process becomes easily parallelizable. Moreover, by conditioning the normalizing flow, the sampling of configurations can be steered towards the regions of interest. We show that this allows for resolving both the thermodynamics and kinetics of the transition region.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge