Ron Stewart
Morgridge Institute for Research, Madison, WI, USA
CHAMMI-75: pre-training multi-channel models with heterogeneous microscopy images
Dec 23, 2025Abstract:Quantifying cell morphology using images and machine learning has proven to be a powerful tool to study the response of cells to treatments. However, models used to quantify cellular morphology are typically trained with a single microscopy imaging type. This results in specialized models that cannot be reused across biological studies because the technical specifications do not match (e.g., different number of channels), or because the target experimental conditions are out of distribution. Here, we present CHAMMI-75, an open access dataset of heterogeneous, multi-channel microscopy images from 75 diverse biological studies. We curated this resource from publicly available sources to investigate cellular morphology models that are channel-adaptive and can process any microscopy image type. Our experiments show that training with CHAMMI-75 can improve performance in multi-channel bioimaging tasks primarily because of its high diversity in microscopy modalities. This work paves the way to create the next generation of cellular morphology models for biological studies.
Machine Learning to Predict Developmental Neurotoxicity with High-throughput Data from 2D Bio-engineered Tissues
May 06, 2019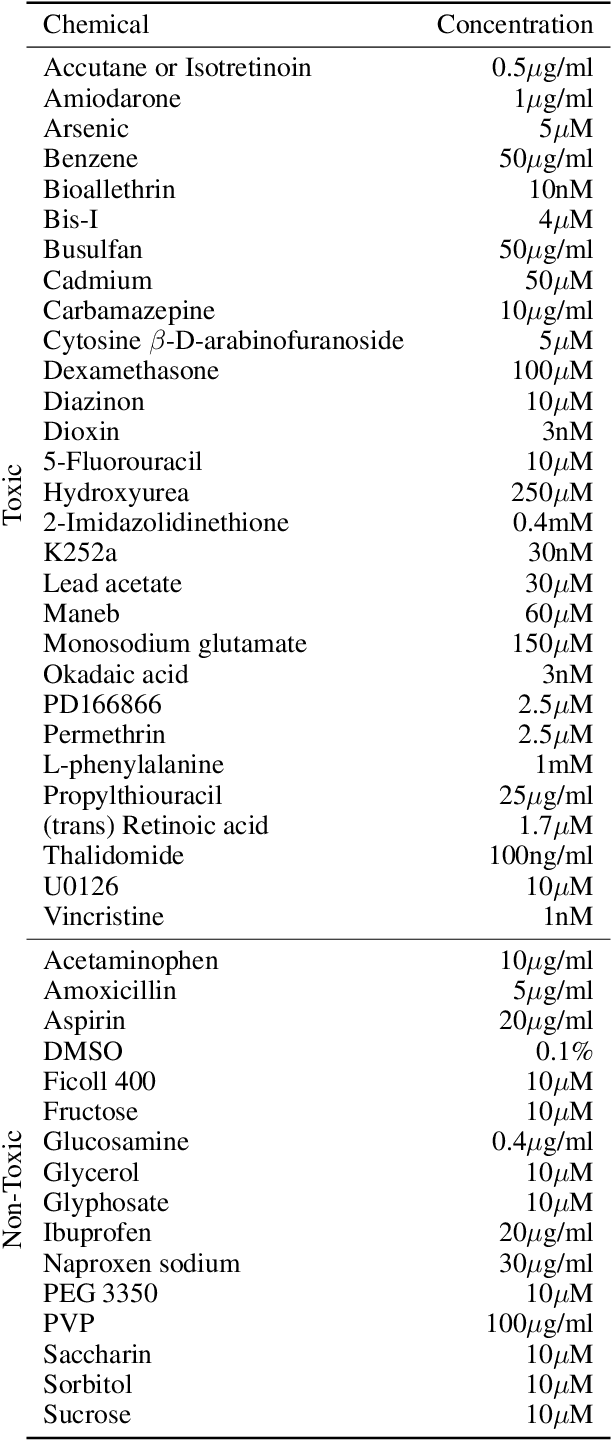
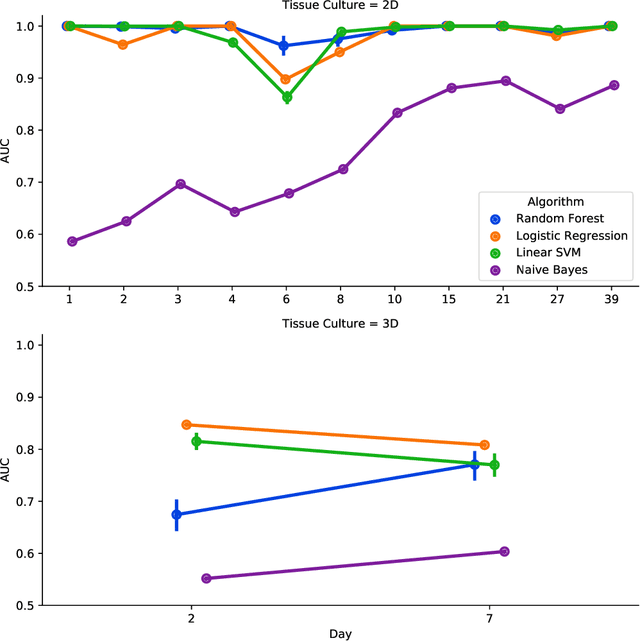
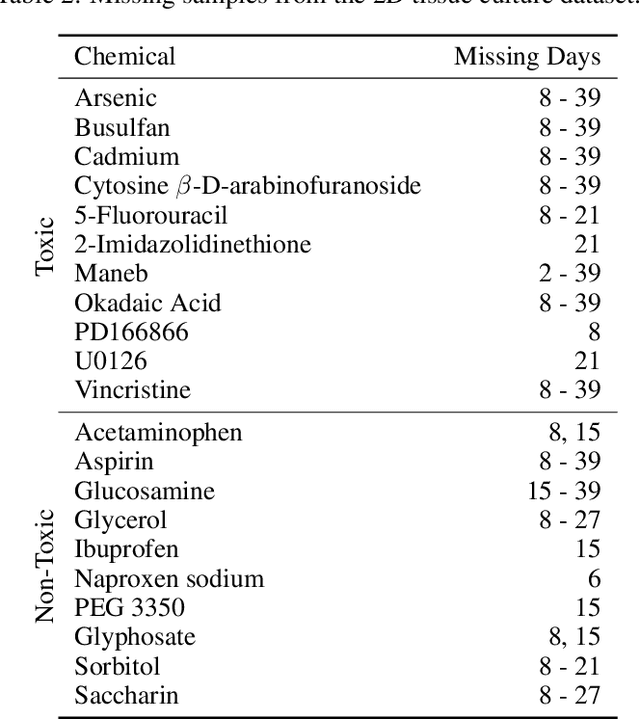
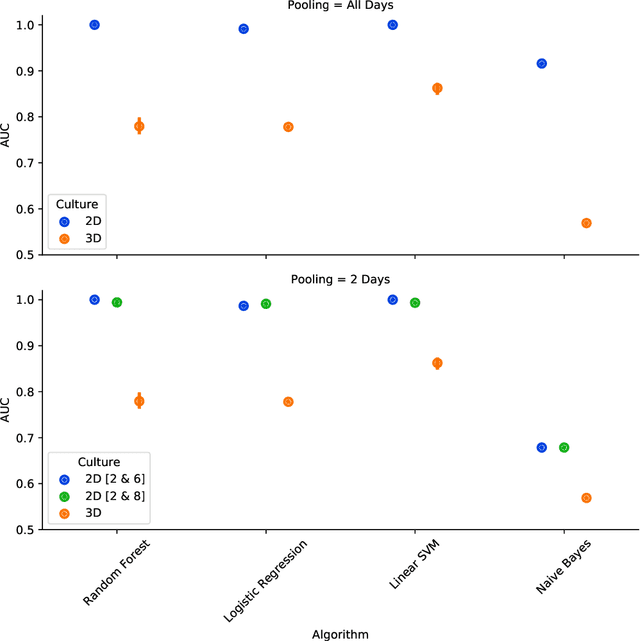
Abstract:There is a growing need for fast and accurate methods for testing developmental neurotoxicity across several chemical exposure sources. Current approaches, such as in vivo animal studies, and assays of animal and human primary cell cultures, suffer from challenges related to time, cost, and applicability to human physiology. We previously demonstrated success employing machine learning to predict developmental neurotoxicity using gene expression data collected from human 3D tissue models exposed to various compounds. The 3D model is biologically similar to developing neural structures, but its complexity necessitates extensive expertise and effort to employ. By instead focusing solely on constructing an assay of developmental neurotoxicity, we propose that a simpler 2D tissue model may prove sufficient. We thus compare the accuracy of predictive models trained on data from a 2D tissue model with those trained on data from a 3D tissue model, and find the 2D model to be substantially more accurate. Furthermore, we find the 2D model to be more robust under stringent gene set selection, whereas the 3D model suffers substantial accuracy degradation. While both approaches have advantages and disadvantages, we propose that our described 2D approach could be a valuable tool for decision makers when prioritizing neurotoxicity screening.
Computational Drug Repositioning Using Continuous Self-controlled Case Series
Apr 20, 2016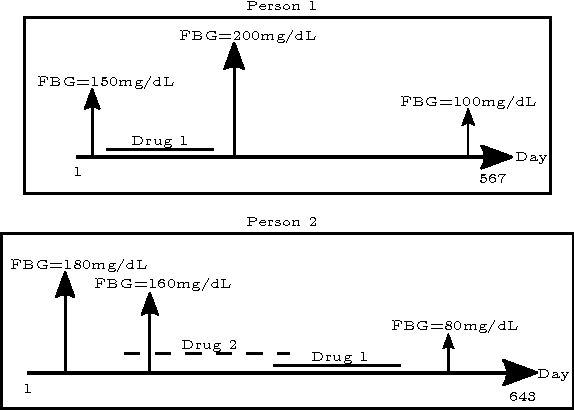
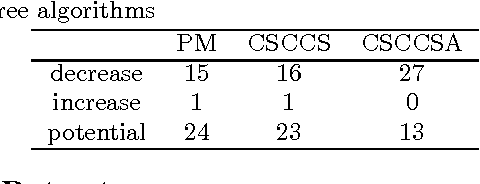
Abstract:Computational Drug Repositioning (CDR) is the task of discovering potential new indications for existing drugs by mining large-scale heterogeneous drug-related data sources. Leveraging the patient-level temporal ordering information between numeric physiological measurements and various drug prescriptions provided in Electronic Health Records (EHRs), we propose a Continuous Self-controlled Case Series (CSCCS) model for CDR. As an initial evaluation, we look for drugs that can control Fasting Blood Glucose (FBG) level in our experiments. Applying CSCCS to the Marshfield Clinic EHR, well-known drugs that are indicated for controlling blood glucose level are rediscovered. Furthermore, some drugs with recent literature support for the potential effect of blood glucose level control are also identified.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge