Rishal Aggarwal
BoltzNCE: Learning Likelihoods for Boltzmann Generation with Stochastic Interpolants and Noise Contrastive Estimation
Jul 02, 2025Abstract:Efficient sampling from the Boltzmann distribution defined by an energy function is a key challenge in modeling physical systems such as molecules. Boltzmann Generators tackle this by leveraging Continuous Normalizing Flows that transform a simple prior into a distribution that can be reweighted to match the Boltzmann distribution using sample likelihoods. However, obtaining likelihoods requires computing costly Jacobians during integration, making it impractical for large molecular systems. To overcome this, we propose learning the likelihood of the generated distribution via an energy-based model trained with noise contrastive estimation and score matching. By using stochastic interpolants to anneal between the prior and generated distributions, we combine both the objective functions to efficiently learn the density function. On the alanine dipeptide system, we demonstrate that our method yields free energy profiles and energy distributions comparable to those obtained with exact likelihoods. Additionally, we show that free energy differences between metastable states can be estimated accurately with orders-of-magnitude speedup.
APObind: A Dataset of Ligand Unbound Protein Conformations for Machine Learning Applications in De Novo Drug Design
Aug 25, 2021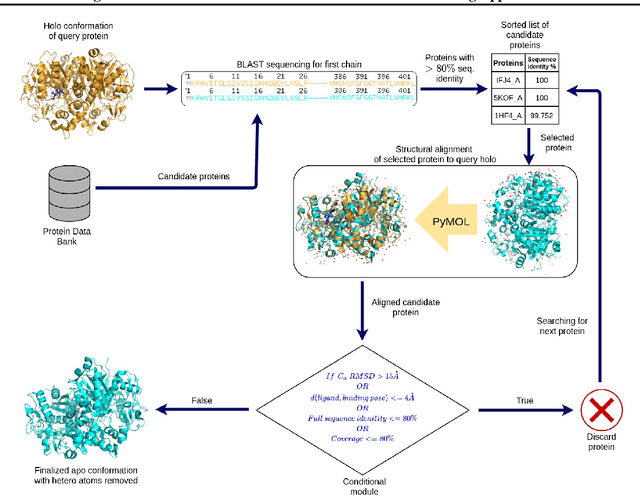

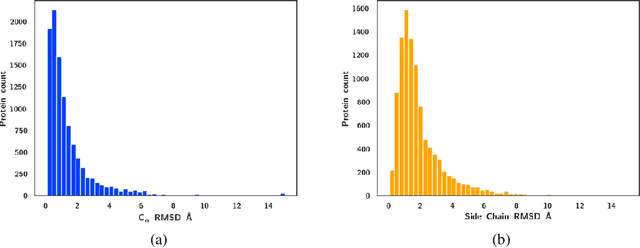
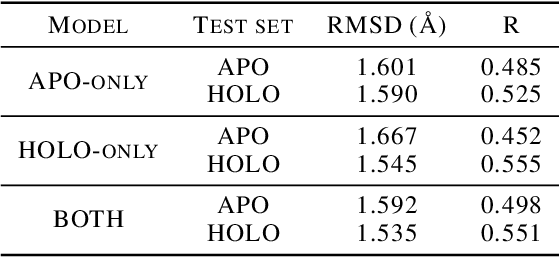
Abstract:Protein-ligand complex structures have been utilised to design benchmark machine learning methods that perform important tasks related to drug design such as receptor binding site detection, small molecule docking and binding affinity prediction. However, these methods are usually trained on only ligand bound (or holo) conformations of the protein and therefore are not guaranteed to perform well when the protein structure is in its native unbound conformation (or apo), which is usually the conformation available for a newly identified receptor. A primary reason for this is that the local structure of the binding site usually changes upon ligand binding. To facilitate solutions for this problem, we propose a dataset called APObind that aims to provide apo conformations of proteins present in the PDBbind dataset, a popular dataset used in drug design. Furthermore, we explore the performance of methods specific to three use cases on this dataset, through which, the importance of validating them on the APObind dataset is demonstrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge