Paul Tafforeau
Multispectral CT Denoising via Simulation-Trained Deep Learning: Experimental Results at the ESRF BM18
Sep 10, 2025Abstract:Multispectral computed tomography (CT) enables advanced material characterization by acquiring energy-resolved projection data. However, since the incoming X-ray flux is be distributed across multiple narrow energy bins, the photon count per bin is greatly reduced compared to standard energy-integrated imaging. This inevitably introduces substantial noise, which can either prolong acquisition times and make scan durations infeasible or degrade image quality with strong noise artifacts. To address this challenge, we present a dedicated neural network-based denoising approach tailored for multispectral CT projections acquired at the BM18 beamline of the ESRF. The method exploits redundancies across angular, spatial, and spectral domains through specialized sub-networks combined via stacked generalization and an attention mechanism. Non-local similarities in the angular-spatial domain are leveraged alongside correlations between adjacent energy bands in the spectral domain, enabling robust noise suppression while preserving fine structural details. Training was performed exclusively on simulated data replicating the physical and noise characteristics of the BM18 setup, with validation conducted on CT scans of custom-designed phantoms containing both high-Z and low-Z materials. The denoised projections and reconstructions demonstrate substantial improvements in image quality compared to classical denoising methods and baseline CNN models. Quantitative evaluations confirm that the proposed method achieves superior performance across a broad spectral range, generalizing effectively to real-world experimental data while significantly reducing noise without compromising structural fidelity.
Deep Learning for Vascular Segmentation and Applications in Phase Contrast Tomography Imaging
Nov 22, 2023Abstract:Automated blood vessel segmentation is vital for biomedical imaging, as vessel changes indicate many pathologies. Still, precise segmentation is difficult due to the complexity of vascular structures, anatomical variations across patients, the scarcity of annotated public datasets, and the quality of images. We present a thorough literature review, highlighting the state of machine learning techniques across diverse organs. Our goal is to provide a foundation on the topic and identify a robust baseline model for application to vascular segmentation in a new imaging modality, Hierarchical Phase Contrast Tomography (HiP CT). Introduced in 2020 at the European Synchrotron Radiation Facility, HiP CT enables 3D imaging of complete organs at an unprecedented resolution of ca. 20mm per voxel, with the capability for localized zooms in selected regions down to 1mm per voxel without sectioning. We have created a training dataset with double annotator validated vascular data from three kidneys imaged with HiP CT in the context of the Human Organ Atlas Project. Finally, utilising the nnU Net model, we conduct experiments to assess the models performance on both familiar and unseen samples, employing vessel specific metrics. Our results show that while segmentations yielded reasonably high scores such as clDice values ranging from 0.82 to 0.88, certain errors persisted. Large vessels that collapsed due to the lack of hydrostatic pressure (HiP CT is an ex vivo technique) were segmented poorly. Moreover, decreased connectivity in finer vessels and higher segmentation errors at vessel boundaries were observed. Such errors obstruct the understanding of the structures by interrupting vascular tree connectivity. Through our review and outputs, we aim to set a benchmark for subsequent model evaluations using various modalities, especially with the HiP CT imaging database.
Automated segmentation of microtomography imaging of Egyptian mummies
May 14, 2021
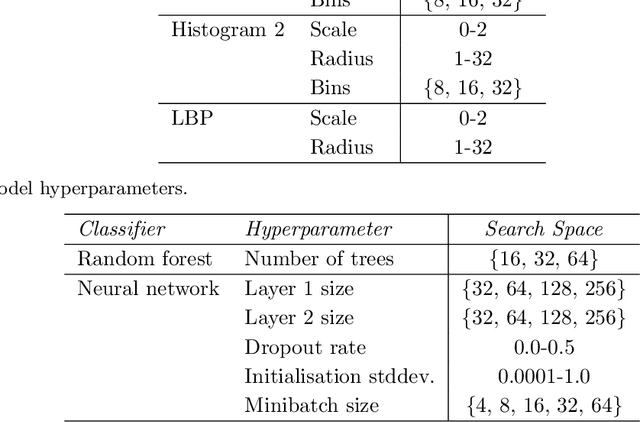
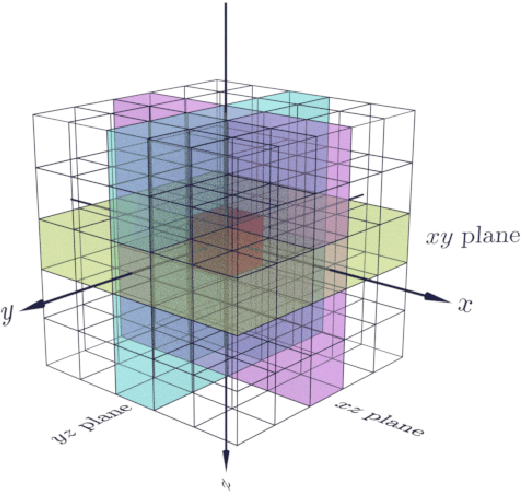
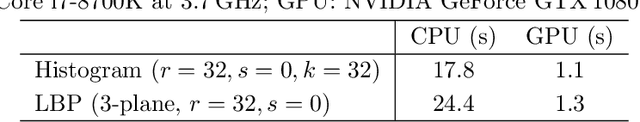
Abstract:Propagation Phase Contrast Synchrotron Microtomography (PPC-SR${\mu}$CT) is the gold standard for non-invasive and non-destructive access to internal structures of archaeological remains. In this analysis, the virtual specimen needs to be segmented to separate different parts or materials, a process that normally requires considerable human effort. In the Automated SEgmentation of Microtomography Imaging (ASEMI) project, we developed a tool to automatically segment these volumetric images, using manually segmented samples to tune and train a machine learning model. For a set of four specimens of ancient Egyptian animal mummies we achieve an overall accuracy of 94-98% when compared with manually segmented slices, approaching the results of off-the-shelf commercial software using deep learning (97-99%) at much lower complexity. A qualitative analysis of the segmented output shows that our results are close in term of usability to those from deep learning, justifying the use of these techniques.
PyHST2: an hybrid distributed code for high speed tomographic reconstruction with iterative reconstruction and a priori knowledge capabilities
Jun 06, 2013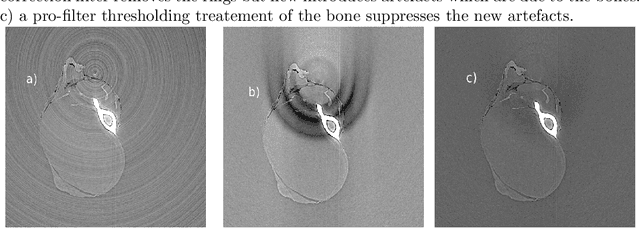
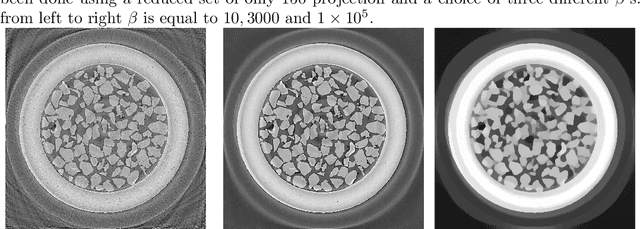
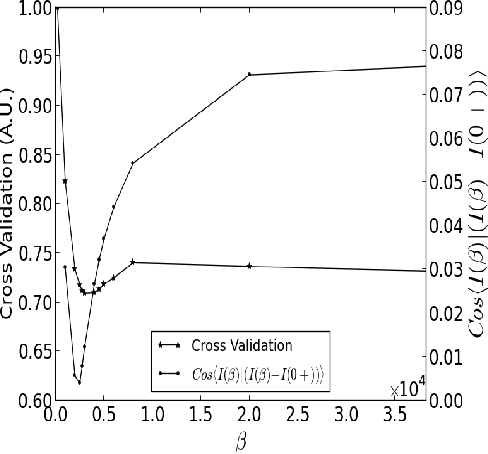
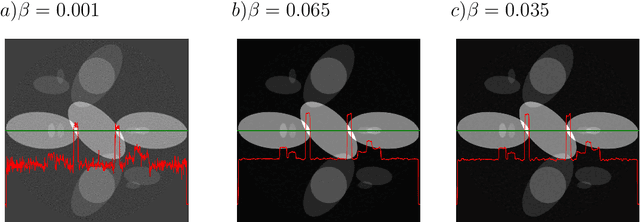
Abstract:We present the PyHST2 code which is in service at ESRF for phase-contrast and absorption tomography. This code has been engineered to sustain the high data flow typical of the third generation synchrotron facilities (10 terabytes per experiment) by adopting a distributed and pipelined architecture. The code implements, beside a default filtered backprojection reconstruction, iterative reconstruction techniques with a-priori knowledge. These latter are used to improve the reconstruction quality or in order to reduce the required data volume and reach a given quality goal. The implemented a-priori knowledge techniques are based on the total variation penalisation and a new recently found convex functional which is based on overlapping patches. We give details of the different methods and their implementations while the code is distributed under free license. We provide methods for estimating, in the absence of ground-truth data, the optimal parameters values for a-priori techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge