Omar S. Al-Kadi
Spatio-Temporal Segmentation in 3D Echocardiographic Sequences using Fractional Brownian Motion
Dec 21, 2019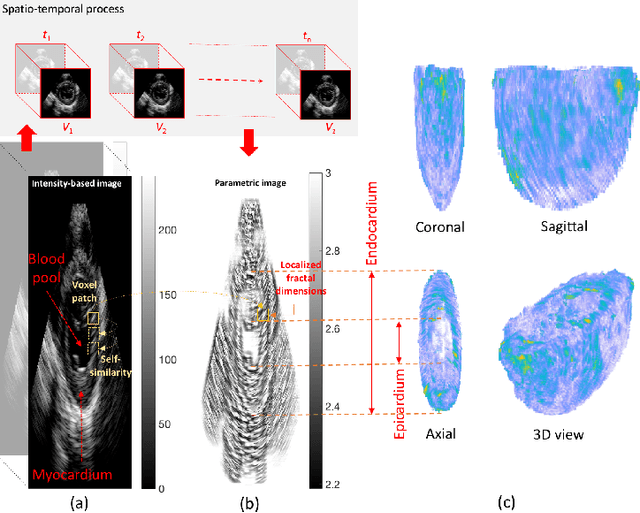
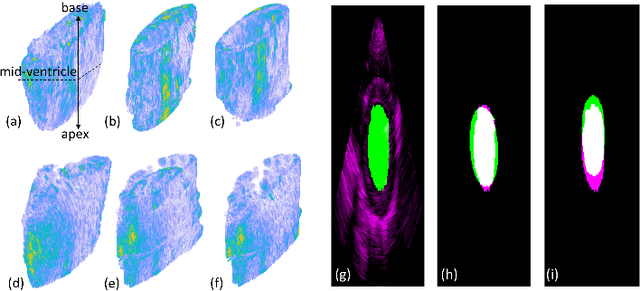
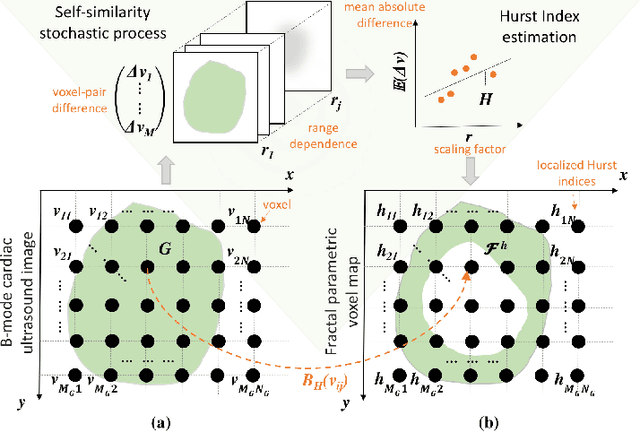
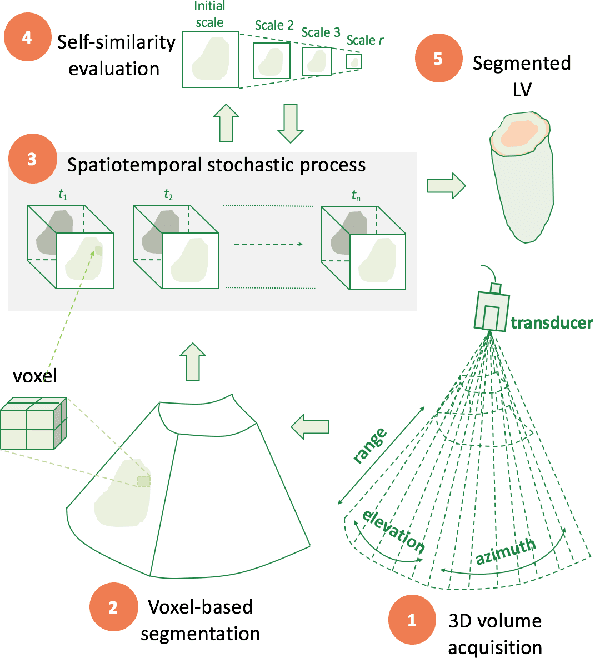
Abstract:An important aspect for an improved cardiac functional analysis is the accurate segmentation of the left ventricle (LV). A novel approach for fully-automated segmentation of the LV endocardium and epicardium contours is presented. This is mainly based on the natural physical characteristics of the LV shape structure. Both sides of the LV boundaries exhibit natural elliptical curvatures by having details on various scales, i.e. exhibiting fractal-like characteristics. The fractional Brownian motion (fBm), which is a non-stationary stochastic process, integrates well with the stochastic nature of ultrasound echoes. It has the advantage of representing a wide range of non-stationary signals and can quantify statistical local self-similarity throughout the time-sequence ultrasound images. The locally characterized boundaries of the fBm segmented LV were further iteratively refined using global information by means of second-order moments. The method is benchmarked using synthetic 3D+time echocardiographic sequences for normal and different ischemic cardiomyopathy, and results compared with state-of-the-art LV segmentation. Furthermore, the framework was validated against real data from canine cases with expert-defined segmentations and demonstrated improved accuracy. The fBm-based segmentation algorithm is fully automatic and has the potential to be used clinically together with 3D echocardiography for improved cardiovascular disease diagnosis.
* 11 pages, 10 figures, 2 tables, journal article
Heterogeneous tissue characterization using ultrasound: a comparison of fractal analysis backscatter models on liver tumors
Dec 20, 2019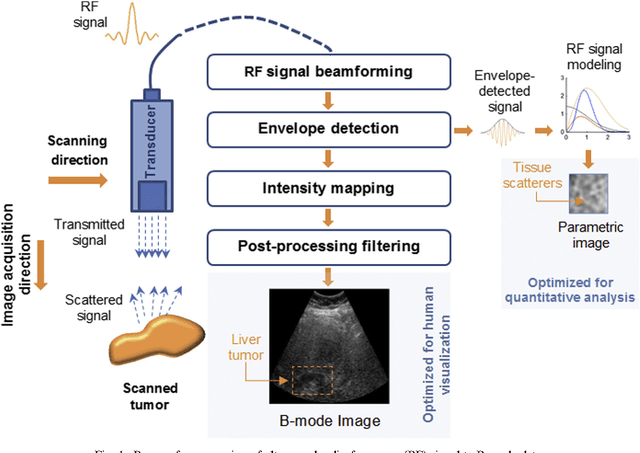
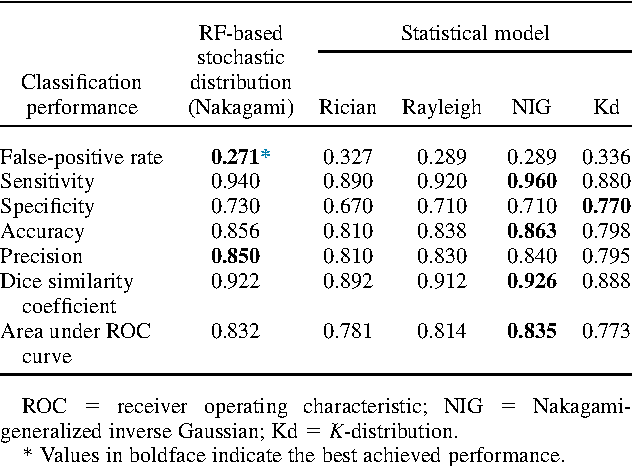
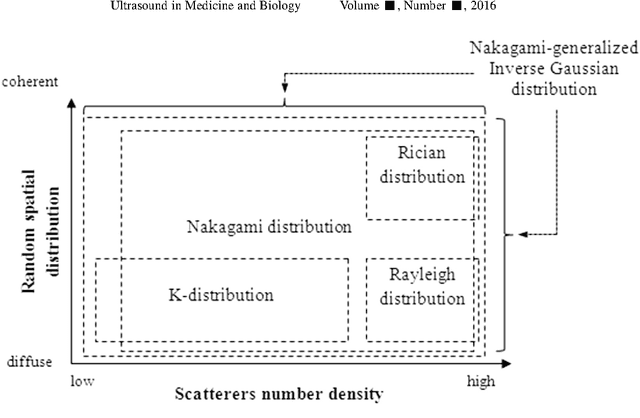
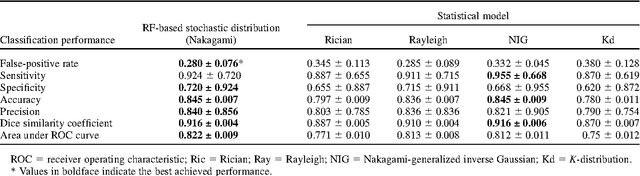
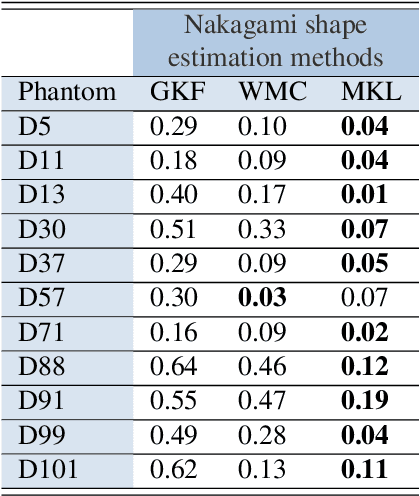
Abstract:Assessing tumor tissue heterogeneity via ultrasound has recently been suggested for predicting early response to treatment. The ultrasound backscattering characteristics can assist in better understanding the tumor texture by highlighting local concentration and spatial arrangement of tissue scatterers. However, it is challenging to quantify the various tissue heterogeneities ranging from fine-to-coarse of the echo envelope peaks in tumor texture. Local parametric fractal features extracted via maximum likelihood estimation from five well-known statistical model families are evaluated for the purpose of ultrasound tissue characterization. The fractal dimension (self-similarity measure) was used to characterize the spatial distribution of scatterers, while the Lacunarity (sparsity measure) was applied to determine scatterer number density. Performance was assessed based on 608 cross-sectional clinical ultrasound RF images of liver tumors (230 and 378 demonstrating respondent and non-respondent cases, respectively). Crossvalidation via leave-one-tumor-out and with different k-folds methodologies using a Bayesian classifier were employed for validation. The fractal properties of the backscattered echoes based on the Nakagami model (Nkg) and its extend four-parameter Nakagami-generalized inverse Gaussian (NIG) distribution achieved best results - with nearly similar performance - for characterizing liver tumor tissue. Accuracy, sensitivity and specificity for the Nkg/NIG were: 85.6%/86.3%, 94.0%/96.0%, and 73.0%/71.0%, respectively. Other statistical models, such as the Rician, Rayleigh, and K-distribution were found to not be as effective in characterizing the subtle changes in tissue texture as an indication of response to treatment. Employing the most relevant and practical statistical model could have potential consequences for the design of an early and effective clinical therapy.
* 31 pages, 7 figures, 3 tables, journal article
Multiscale Nakagami parametric imaging for improved liver tumor localization
Jun 11, 2019
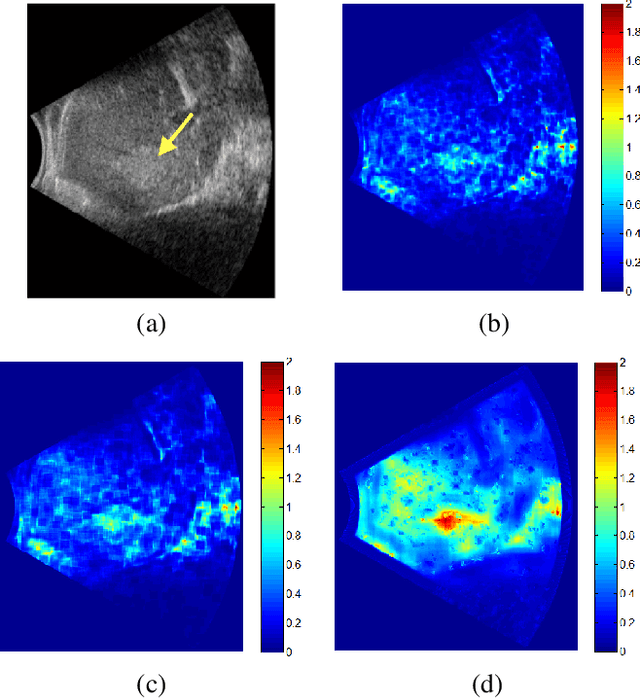
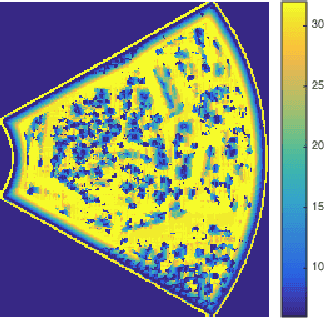
Abstract:Effective ultrasound tissue characterization is usually hindered by complex tissue structures. The interlacing of speckle patterns complicates the correct estimation of backscatter distribution parameters. Nakagami parametric imaging based on localized shape parameter mapping can model different backscattering conditions. However, performance of the constructed Nakagami image depends on the sensitivity of the estimation method to the backscattered statistics and scale of analysis. Using a fixed focal region of interest in estimating the Nakagami parametric image would increase estimation variance. In this work, localized Nakagami parameters are estimated adaptively by means of maximum likelihood estimation on a multiscale basis. The varying size kernel integrates the goodness-of-fit of the backscattering distribution parameters at multiple scales for more stable parameter estimation. Results show improved quantitative visualization of changes in tissue specular reflections, suggesting a potential approach for improving tumor localization in low contrast ultrasound images.
A Gabor Filter Texture Analysis Approach for Histopathological Brain Tumor Subtype Discrimination
Apr 17, 2017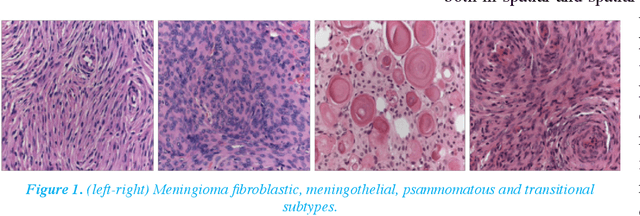
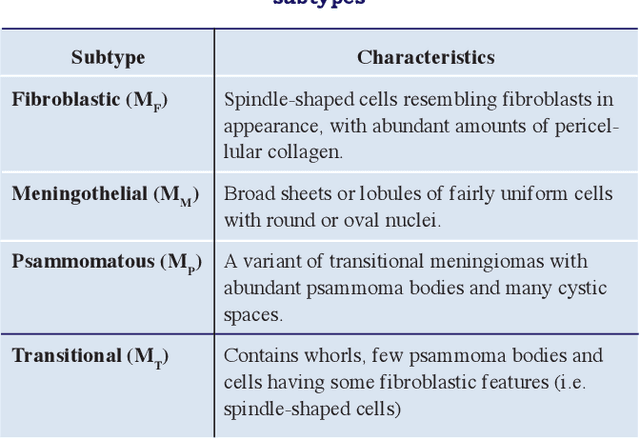
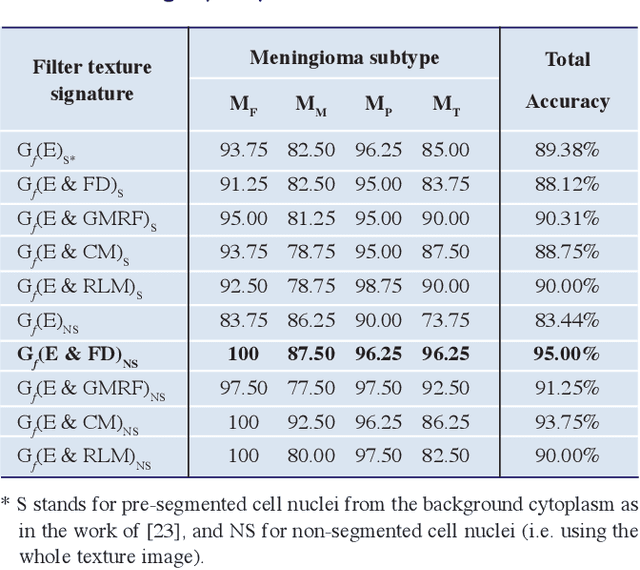
Abstract:Meningioma brain tumour discrimination is challenging as many histological patterns are mixed between the different subtypes. In clinical practice, dominant patterns are investigated for signs of specific meningioma pathology; however the simple observation could result in inter- and intra-observer variation due to the complexity of the histopathological patterns. Also employing a computerised feature extraction approach applied at a single resolution scale might not suffice in accurately delineating the mixture of histopathological patterns. In this work we propose a novel multiresolution feature extraction approach for characterising the textural properties of the different pathological patterns (i.e. mainly cell nuclei shape, orientation and spatial arrangement within the cytoplasm). The pattern textural properties are characterised at various scales and orientations for an improved separability between the different extracted features. The Gabor filter energy output of each magnitude response was combined with four other fixed-resolution texture signatures (2 model-based and 2 statistical-based) with and without cell nuclei segmentation. The highest classification accuracy of 95% was reported when combining the Gabor filters energy and the meningioma subimage fractal signature as a feature vector without performing any prior cell nuceli segmentation. This indicates that characterising the cell-nuclei self-similarity properties via Gabor filters can assists in achieving an improved meningioma subtype classification, which can assist in overcoming variations in reported diagnosis.
* 14 pages,4 figures, 2 tables
Supervised Texture Segmentation: A Comparative Study
Jan 02, 2016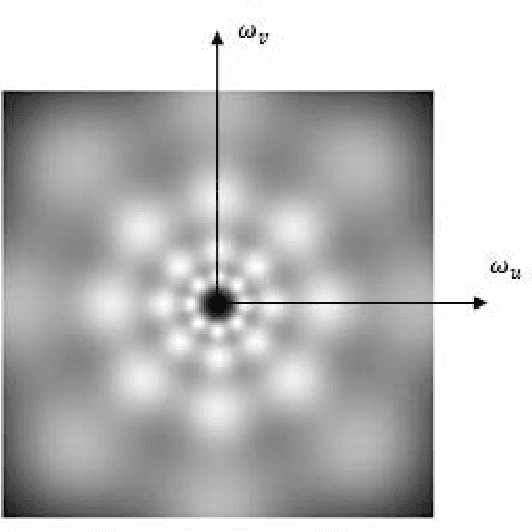
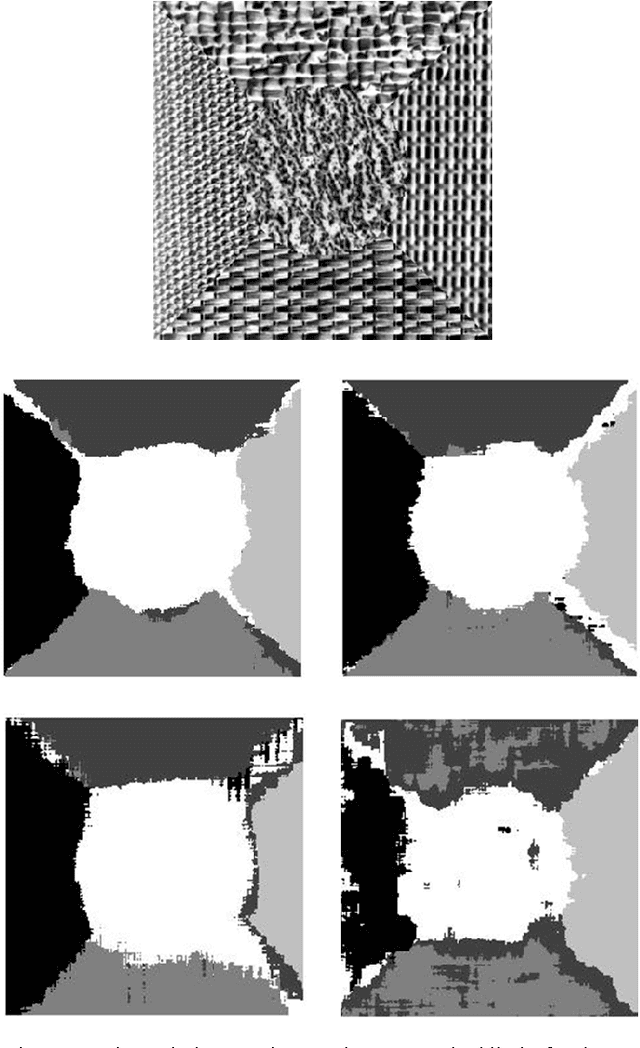

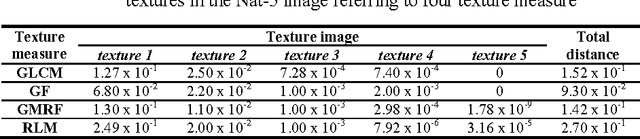
Abstract:This paper aims to compare between four different types of feature extraction approaches in terms of texture segmentation. The feature extraction methods that were used for segmentation are Gabor filters (GF), Gaussian Markov random fields (GMRF), run-length matrix (RLM) and co-occurrence matrix (GLCM). It was shown that the GF performed best in terms of quality of segmentation while the GLCM localises the texture boundaries better as compared to the other methods.
A fractal dimension based optimal wavelet packet analysis technique for classification of meningioma brain tumours
Jan 02, 2016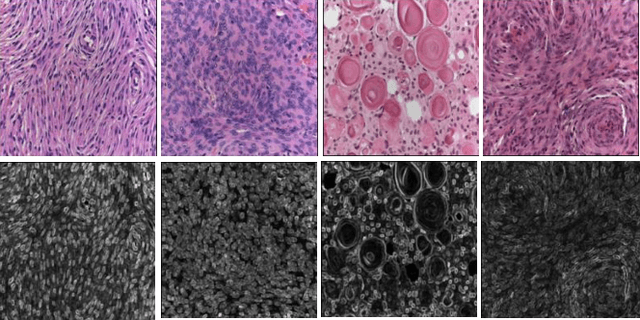
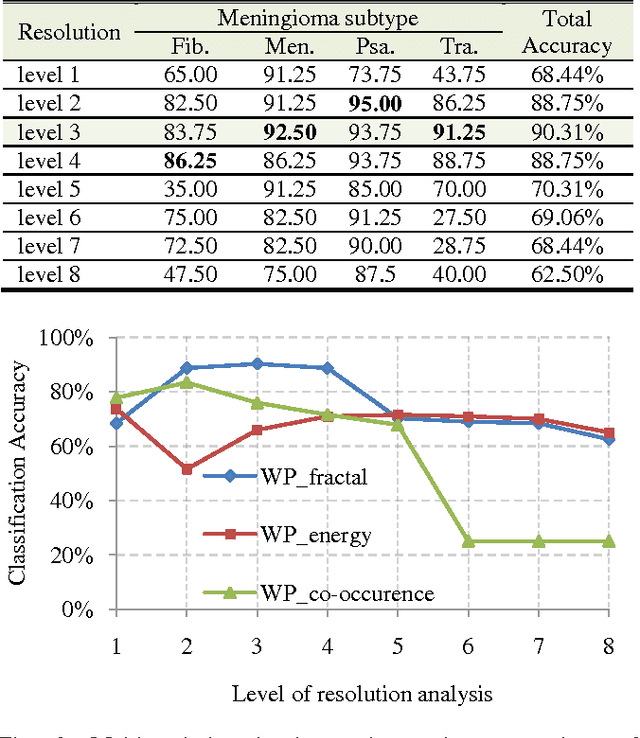
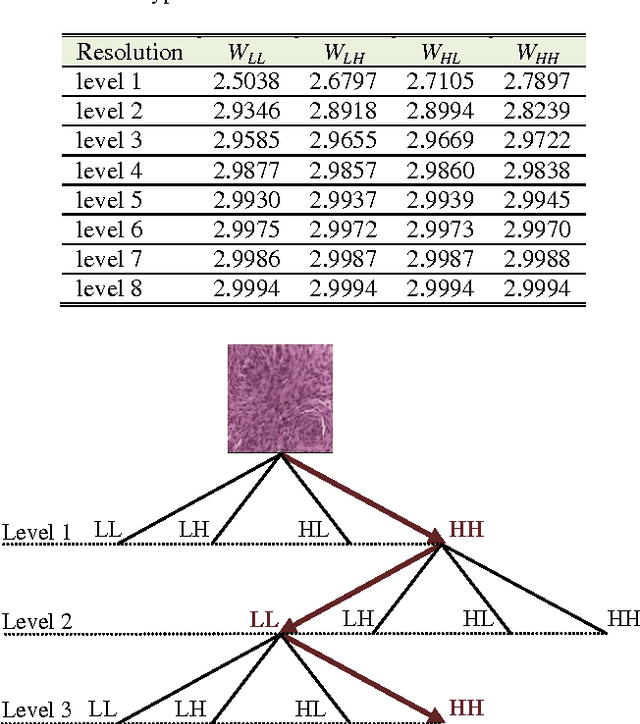
Abstract:With the heterogeneous nature of tissue texture, using a single resolution approach for optimum classification might not suffice. In contrast, a multiresolution wavelet packet analysis can decompose the input signal into a set of frequency subbands giving the opportunity to characterise the texture at the appropriate frequency channel. An adaptive best bases algorithm for optimal bases selection for meningioma histopathological images is proposed, via applying the fractal dimension (FD) as the bases selection criterion in a tree-structured manner. Thereby, the most significant subband that better identifies texture discontinuities will only be chosen for further decomposition, and its fractal signature would represent the extracted feature vector for classification. The best basis selection using the FD outperformed the energy based selection approaches, achieving an overall classification accuracy of 91.25% as compared to 83.44% and 73.75% for the co-occurrence matrix and energy texture signatures; respectively.
A Multiresolution Clinical Decision Support System Based on Fractal Model Design for Classification of Histological Brain Tumours
Dec 25, 2015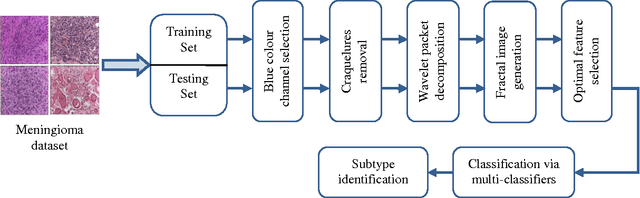
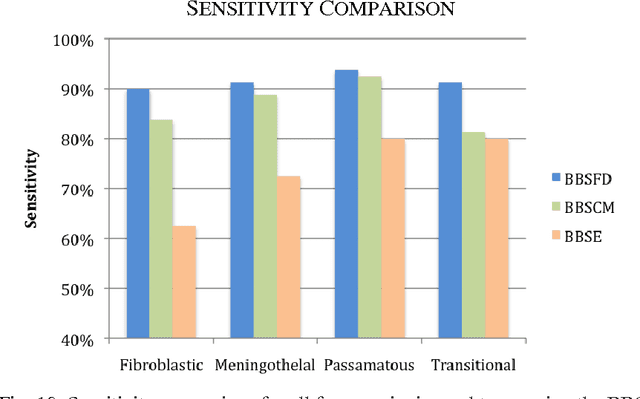
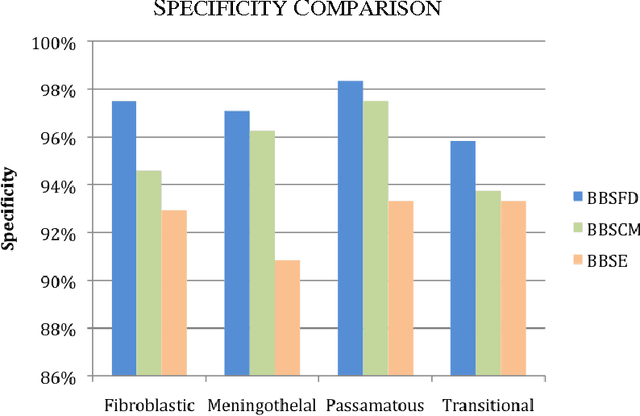
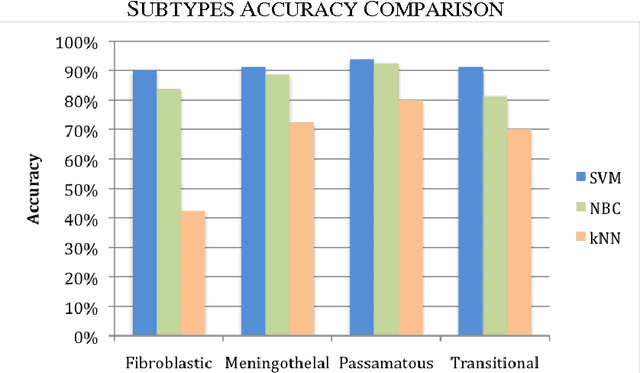
Abstract:Tissue texture is known to exhibit a heterogeneous or non-stationary nature, therefore using a single resolution approach for optimum classification might not suffice. A clinical decision support system that exploits the subband textural fractal characteristics for best bases selection of meningioma brain histopathological image classification is proposed. Each subband is analysed using its fractal dimension instead of energy, which has the advantage of being less sensitive to image intensity and abrupt changes in tissue texture. The most significant subband that best identifies texture discontinuities will be chosen for further decomposition, and its fractal characteristics would represent the optimal feature vector for classification. The performance was tested using the support vector machine (SVM), Bayesian and k-nearest neighbour (kNN) classifiers and a leave-one-patient-out method was employed for validation. Our method outperformed the classical energy based selection approaches, achieving for SVM, Bayesian and kNN classifiers an overall classification accuracy of 94.12%, 92.50% and 79.70%, as compared to 86.31%, 83.19% and 51.63% for the co-occurrence matrix, and 76.01%, 73.50% and 50.69% for the energy texture signatures, respectively. These results indicate the potential usefulness as a decision support system that could complement radiologists diagnostic capability to discriminate higher order statistical textural information, for which it would be otherwise difficult via ordinary human vision.
Texture measures combination for improved meningioma classification of histopathological images
Dec 25, 2015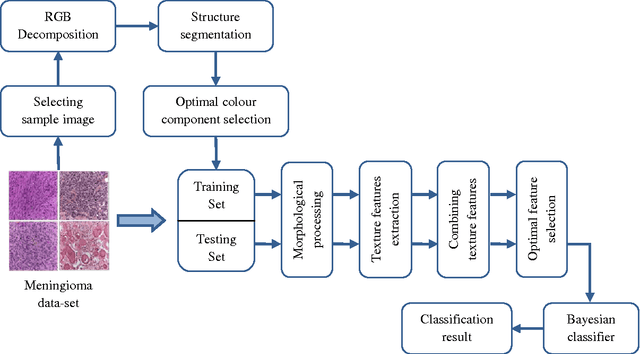
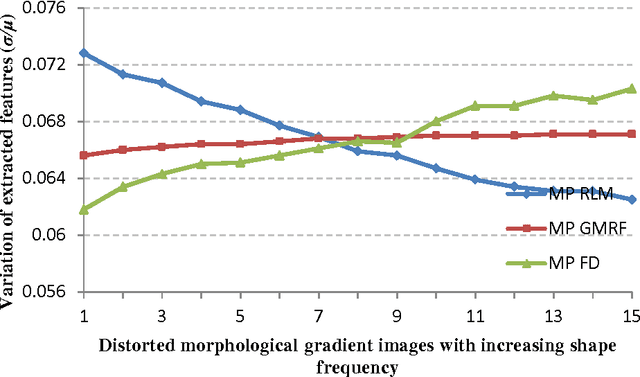
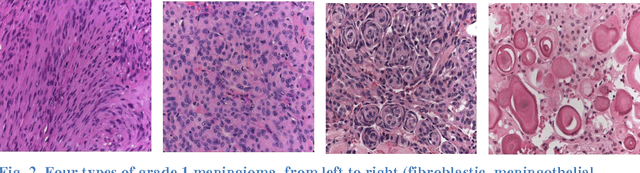
Abstract:Providing an improved technique which can assist pathologists in correctly classifying meningioma tumours with a significant accuracy is our main objective. The proposed technique, which is based on optimum texture measure combination, inspects the separability of the RGB colour channels and selects the channel which best segments the cell nuclei of the histopathological images. The morphological gradient was applied to extract the region of interest for each subtype and for elimination of possible noise (e.g. cracks) which might occur during biopsy preparation. Meningioma texture features are extracted by four different texture measures (two model-based and two statistical-based) and then corresponding features are fused together in different combinations after excluding highly correlated features, and a Bayesian classifier was used for meningioma subtype discrimination. The combined Gaussian Markov random field and run-length matrix texture measures outperformed all other combinations in terms of quantitatively characterising the meningioma tissue, achieving an overall classification accuracy of 92.50%, improving from 83.75% which is the best accuracy achieved if the texture measures are used individually.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge