Ola Spjuth
AI Agents in Drug Discovery
Oct 31, 2025Abstract:Artificial intelligence (AI) agents are emerging as transformative tools in drug discovery, with the ability to autonomously reason, act, and learn through complicated research workflows. Building on large language models (LLMs) coupled with perception, computation, action, and memory tools, these agentic AI systems could integrate diverse biomedical data, execute tasks, carry out experiments via robotic platforms, and iteratively refine hypotheses in closed loops. We provide a conceptual and technical overview of agentic AI architectures, ranging from ReAct and Reflection to Supervisor and Swarm systems, and illustrate their applications across key stages of drug discovery, including literature synthesis, toxicity prediction, automated protocol generation, small-molecule synthesis, drug repurposing, and end-to-end decision-making. To our knowledge, this represents the first comprehensive work to present real-world implementations and quantifiable impacts of agentic AI systems deployed in operational drug discovery settings. Early implementations demonstrate substantial gains in speed, reproducibility, and scalability, compressing workflows that once took months into hours while maintaining scientific traceability. We discuss the current challenges related to data heterogeneity, system reliability, privacy, and benchmarking, and outline future directions towards technology in support of science and translation.
Scalable federated machine learning with FEDn
Feb 27, 2021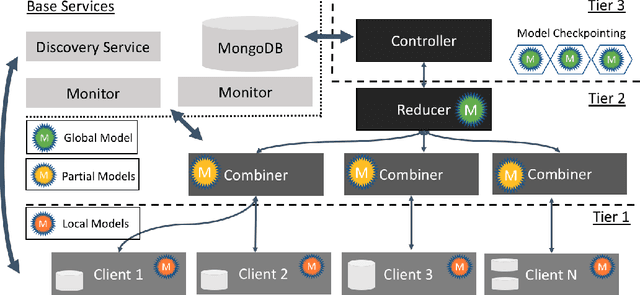

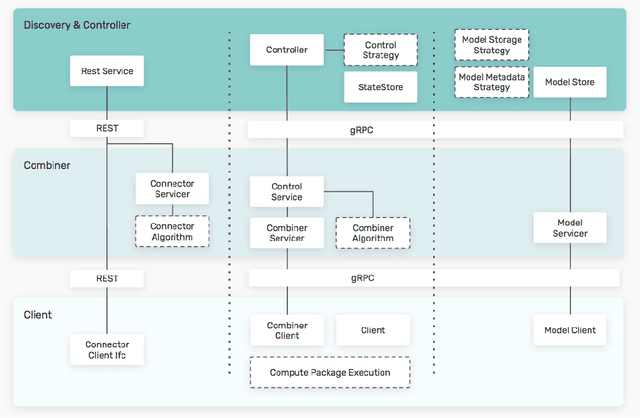
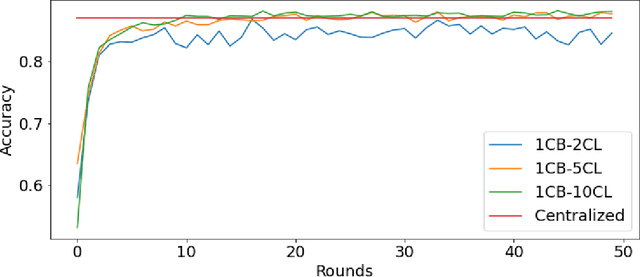
Abstract:Federated machine learning has great promise to overcome the input privacy challenge in machine learning. The appearance of several projects capable of simulating federated learning has led to a corresponding rapid progress on algorithmic aspects of the problem. However, there is still a lack of federated machine learning frameworks that focus on fundamental aspects such as scalability, robustness, security, and performance in a geographically distributed setting. To bridge this gap we have designed and developed the FEDn framework. A main feature of FEDn is to support both cross-device and cross-silo training settings. This makes FEDn a powerful tool for researching a wide range of machine learning applications in a realistic setting.
Combining Prediction Intervals on Multi-Source Non-Disclosed Regression Datasets
Aug 15, 2019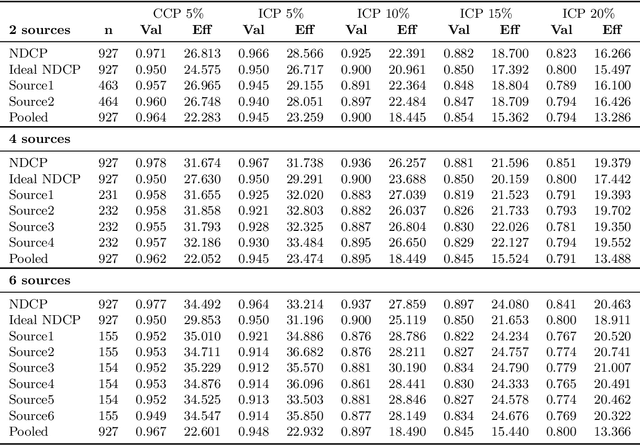
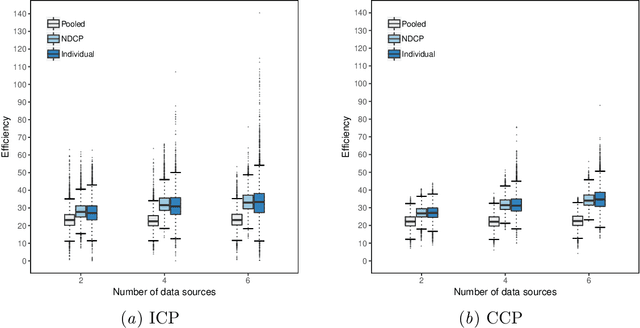
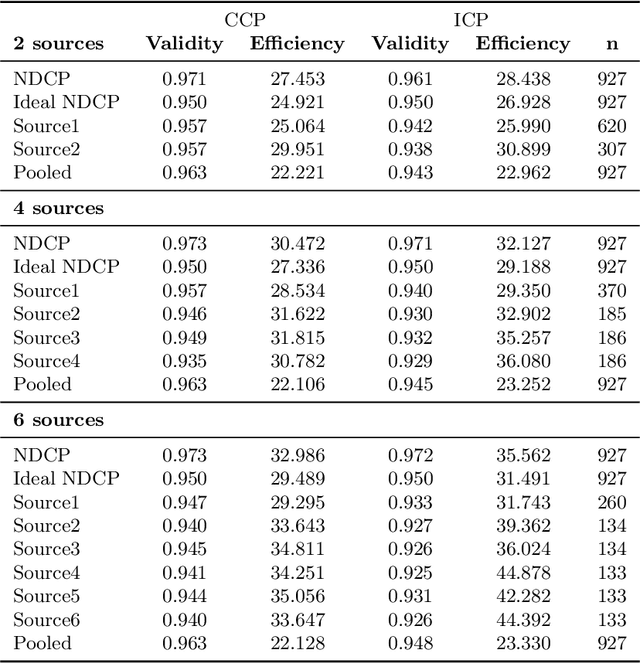
Abstract:Conformal Prediction is a framework that produces prediction intervals based on the output from a machine learning algorithm. In this paper we explore the case when training data is made up of multiple parts available in different sources that cannot be pooled. We here consider the regression case and propose a method where a conformal predictor is trained on each data source independently, and where the prediction intervals are then combined into a single interval. We call the approach Non-Disclosed Conformal Prediction (NDCP), and we evaluate it on a regression dataset from the UCI machine learning repository using support vector regression as the underlying machine learning algorithm, with varying number of data sources and sizes. The results show that the proposed method produces conservatively valid prediction intervals, and while we cannot retain the same efficiency as when all data is used, efficiency is improved through the proposed approach as compared to predicting using a single arbitrarily chosen source.
Aggregating Predictions on Multiple Non-disclosed Datasets using Conformal Prediction
Jun 14, 2018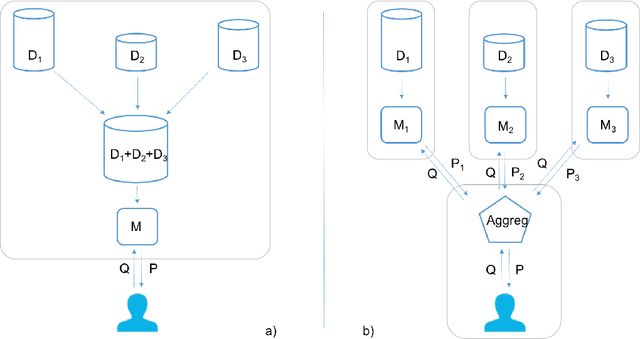
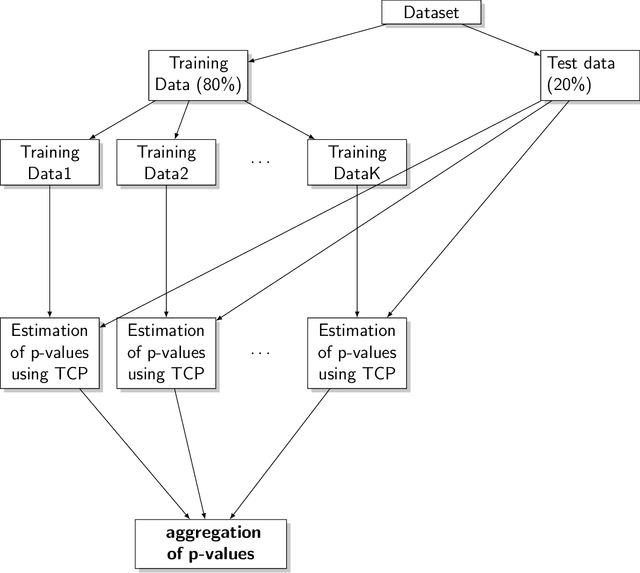
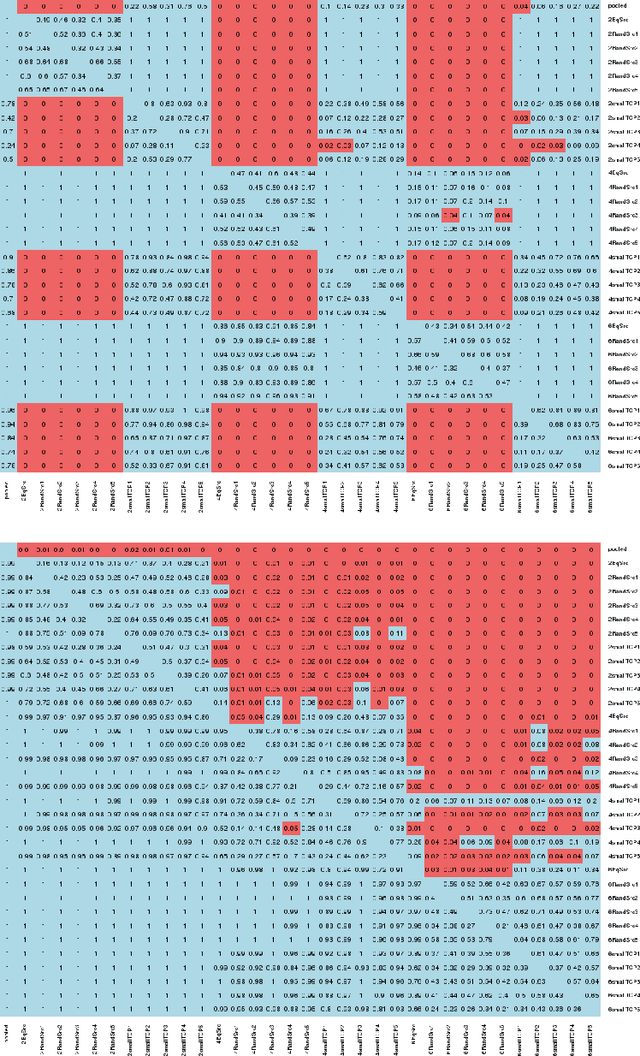
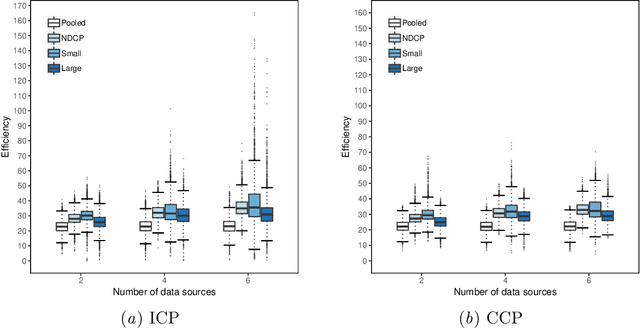
Abstract:Conformal Prediction is a machine learning methodology that produces valid prediction regions under mild conditions. In this paper, we explore the application of making predictions over multiple data sources of different sizes without disclosing data between the sources. We propose that each data source applies a transductive conformal predictor independently using the local data, and that the individual predictions are then aggregated to form a combined prediction region. We demonstrate the method on several data sets, and show that the proposed method produces conservatively valid predictions and reduces the variance in the aggregated predictions. We also study the effect that the number of data sources and size of each source has on aggregated predictions, as compared with equally sized sources and pooled data.
conformalClassification: A Conformal Prediction R Package for Classification
Apr 16, 2018Abstract:The conformalClassification package implements Transductive Conformal Prediction (TCP) and Inductive Conformal Prediction (ICP) for classification problems. Conformal Prediction (CP) is a framework that complements the predictions of machine learning algorithms with reliable measures of confidence. TCP gives results with higher validity than ICP, however ICP is computationally faster than TCP. The package conformalClassification is built upon the random forest method, where votes of the random forest for each class are considered as the conformity scores for each data point. Although the main aim of the conformalClassification package is to generate CP errors (p-values) for classification problems, the package also implements various diagnostic measures such as deviation from validity, error rate, efficiency, observed fuzziness and calibration plots. In future releases, we plan to extend the package to use other machine learning algorithms, (e.g. support vector machines) for model fitting.
Conformal Prediction in Learning Under Privileged Information Paradigm with Applications in Drug Discovery
Apr 04, 2018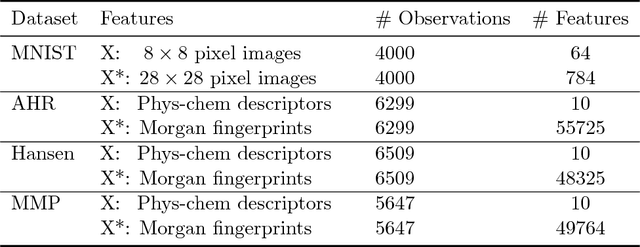
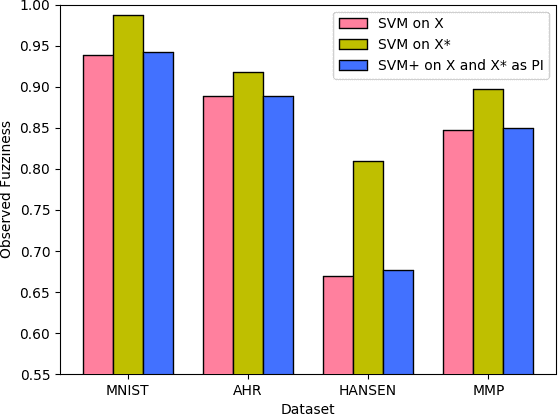
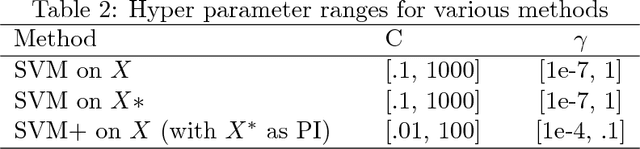
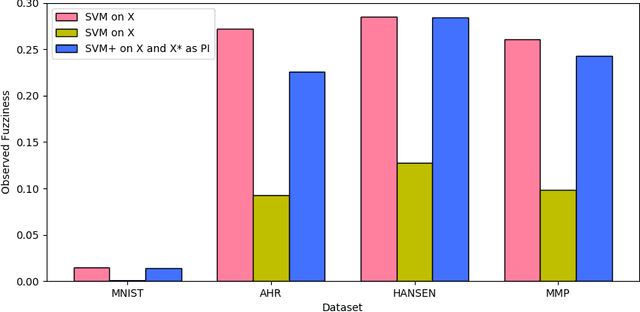
Abstract:This paper explores conformal prediction in the learning under privileged information (LUPI) paradigm. We use the SVM+ realization of LUPI in an inductive conformal predictor, and apply it to the MNIST benchmark dataset and three datasets in drug discovery. The results show that using privileged information produces valid models and improves efficiency compared to standard SVM, however the improvement varies between the tested datasets and is not substantial in the drug discovery applications. More importantly, using SVM+ in a conformal prediction framework enables valid prediction intervals at specified significance levels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge