Ola Engkvist
Parallel Capsule Networks for Classification of White Blood Cells
Sep 06, 2021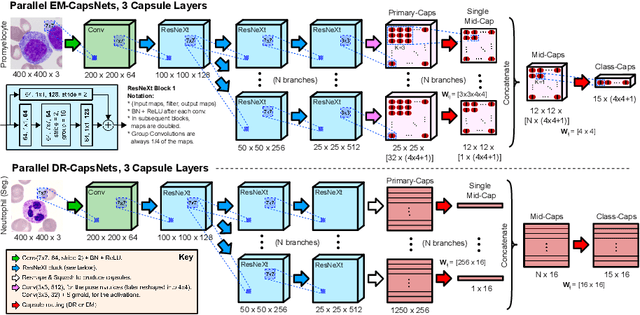
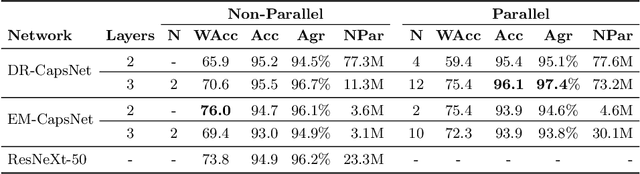
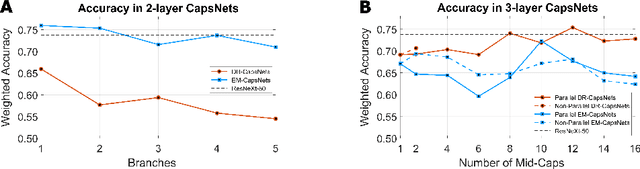
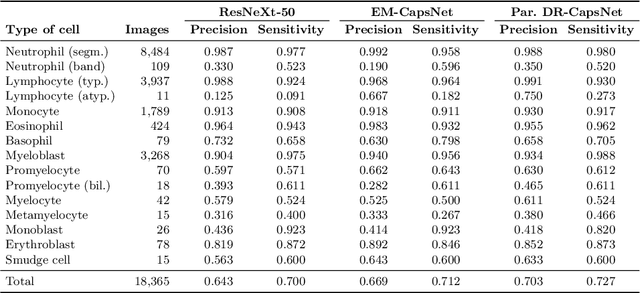
Abstract:Capsule Networks (CapsNets) is a machine learning architecture proposed to overcome some of the shortcomings of convolutional neural networks (CNNs). However, CapsNets have mainly outperformed CNNs in datasets where images are small and/or the objects to identify have minimal background noise. In this work, we present a new architecture, parallel CapsNets, which exploits the concept of branching the network to isolate certain capsules, allowing each branch to identify different entities. We applied our concept to the two current types of CapsNet architectures, studying the performance for networks with different layers of capsules. We tested our design in a public, highly unbalanced dataset of acute myeloid leukaemia images (15 classes). Our experiments showed that conventional CapsNets show similar performance than our baseline CNN (ResNeXt-50) but depict instability problems. In contrast, parallel CapsNets can outperform ResNeXt-50, is more stable, and shows better rotational invariance than both, conventional CapsNets and ResNeXt-50.
Understanding the Performance of Knowledge Graph Embeddings in Drug Discovery
Jun 07, 2021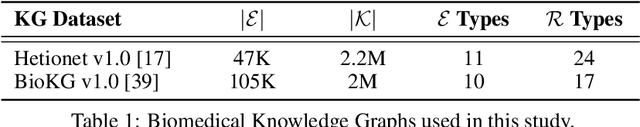
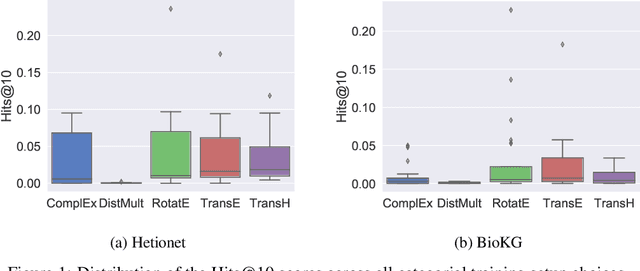
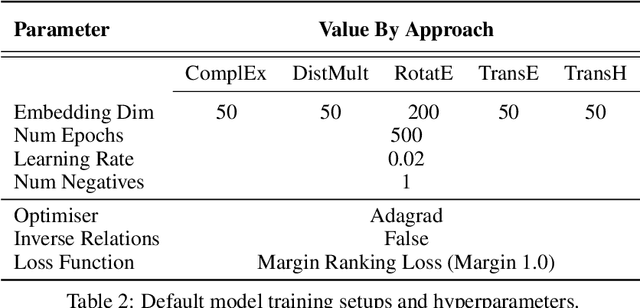
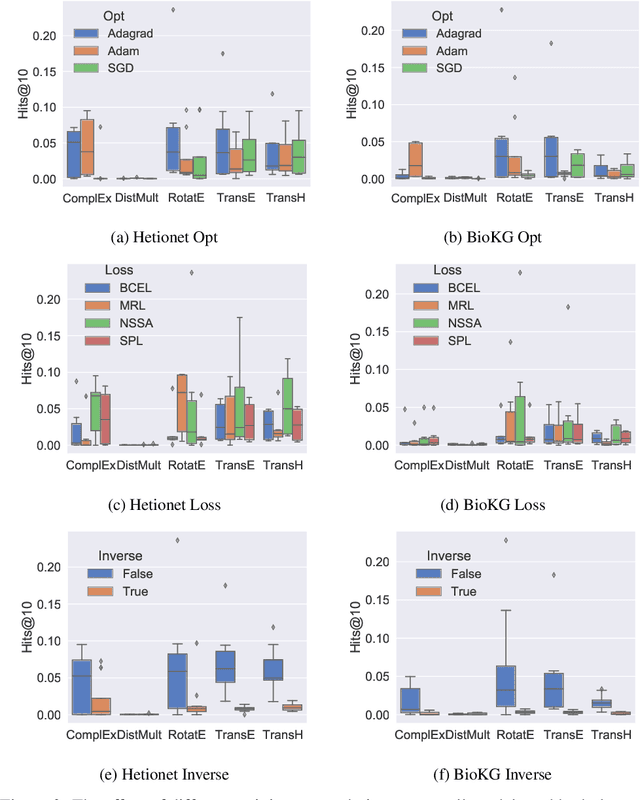
Abstract:Knowledge Graphs (KG) and associated Knowledge Graph Embedding (KGE) models have recently begun to be explored in the context of drug discovery and have the potential to assist in key challenges such as target identification. In the drug discovery domain, KGs can be employed as part of a process which can result in lab-based experiments being performed, or impact on other decisions, incurring significant time and financial costs and most importantly, ultimately influencing patient healthcare. For KGE models to have impact in this domain, a better understanding of not only of performance, but also the various factors which determine it, is required. In this study we investigate, over the course of many thousands of experiments, the predictive performance of five KGE models on two public drug discovery-oriented KGs. Our goal is not to focus on the best overall model or configuration, instead we take a deeper look at how performance can be affected by changes in the training setup, choice of hyperparameters, model parameter initialisation seed and different splits of the datasets. Our results highlight that these factors have significant impact on performance and can even affect the ranking of models. Indeed these factors should be reported along with model architectures to ensure complete reproducibility and fair comparisons of future work, and we argue this is critical for the acceptance of use, and impact of KGEs in a biomedical setting. To aid reproducibility of our own work, we release all experimentation code.
A Review of Biomedical Datasets Relating to Drug Discovery: A Knowledge Graph Perspective
Feb 26, 2021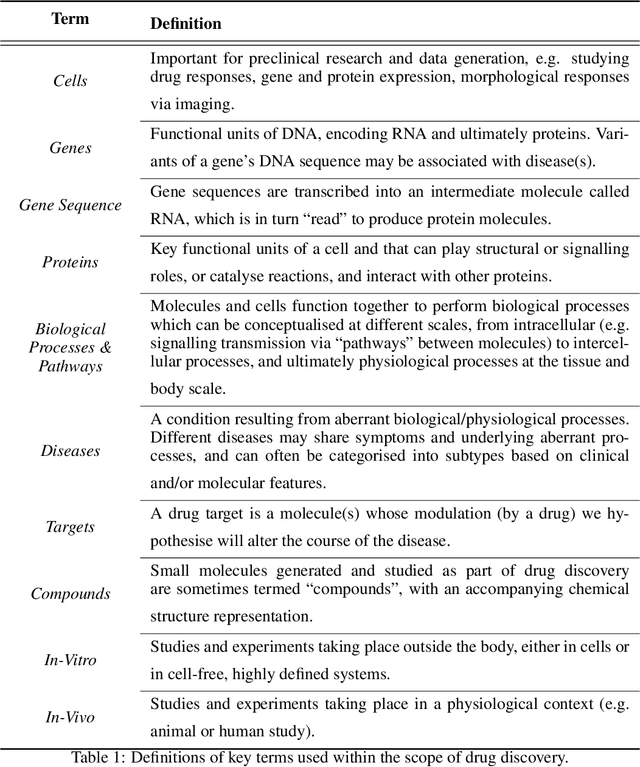
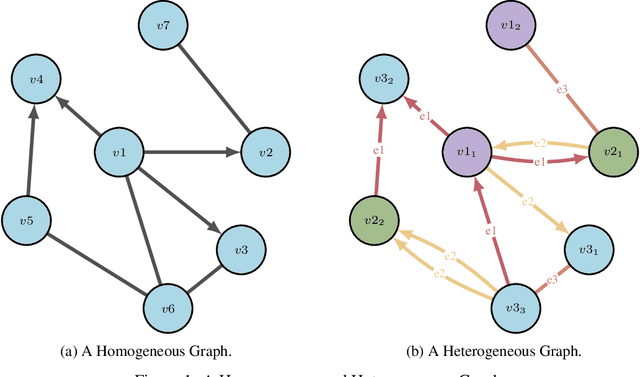
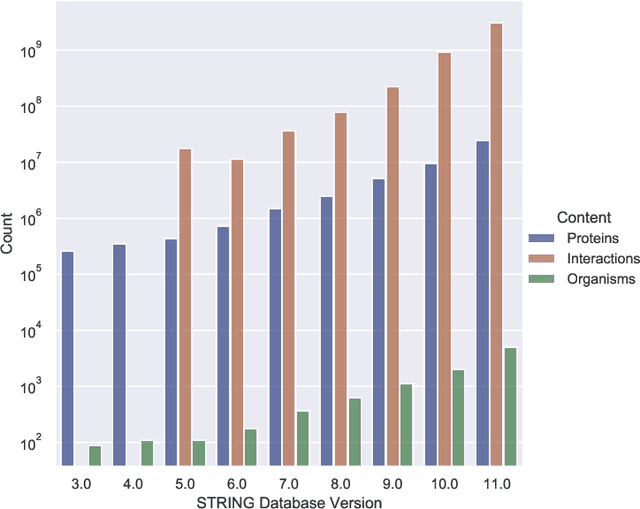
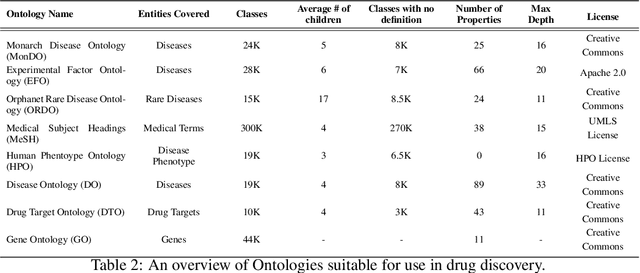
Abstract:Drug discovery and development is an extremely complex process, with high attrition contributing to the costs of delivering new medicines to patients. Recently, various machine learning approaches have been proposed and investigated to help improve the effectiveness and speed of multiple stages of the drug discovery pipeline. Among these techniques, it is especially those using Knowledge Graphs that are proving to have considerable promise across a range of tasks, including drug repurposing, drug toxicity prediction and target gene-disease prioritisation. In such a knowledge graph-based representation of drug discovery domains, crucial elements including genes, diseases and drugs are represented as entities or vertices, whilst relationships or edges between them indicate some level of interaction. For example, an edge between a disease and drug entity might represent a successful clinical trial, or an edge between two drug entities could indicate a potentially harmful interaction. In order to construct high-quality and ultimately informative knowledge graphs however, suitable data and information is of course required. In this review, we detail publicly available primary data sources containing information suitable for use in constructing various drug discovery focused knowledge graphs. We aim to help guide machine learning and knowledge graph practitioners who are interested in applying new techniques to the drug discovery field, but who may be unfamiliar with the relevant data sources. Overall we hope this review will help motivate more machine learning researchers to explore combining knowledge graphs and machine learning to help solve key and emerging questions in the drug discovery domain.
Application of generative autoencoder in de novo molecular design
Nov 21, 2017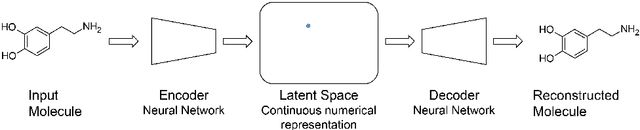
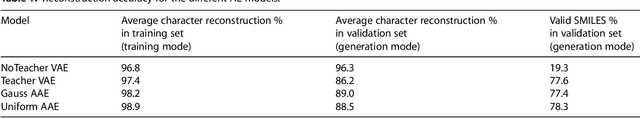
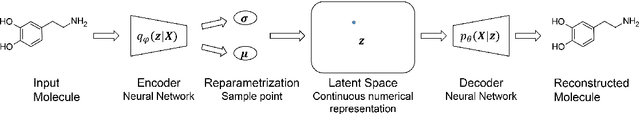

Abstract:A major challenge in computational chemistry is the generation of novel molecular structures with desirable pharmacological and physiochemical properties. In this work, we investigate the potential use of autoencoder, a deep learning methodology, for de novo molecular design. Various generative autoencoders were used to map molecule structures into a continuous latent space and vice versa and their performance as structure generator was assessed. Our results show that the latent space preserves chemical similarity principle and thus can be used for the generation of analogue structures. Furthermore, the latent space created by autoencoders were searched systematically to generate novel compounds with predicted activity against dopamine receptor type 2 and compounds similar to known active compounds not included in the training set were identified.
Molecular De Novo Design through Deep Reinforcement Learning
Aug 29, 2017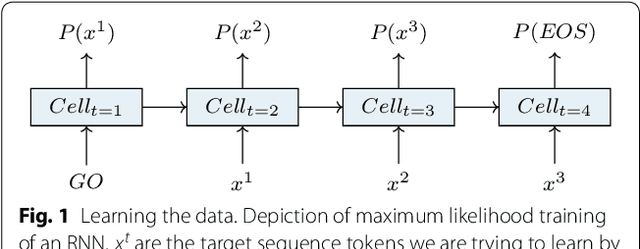
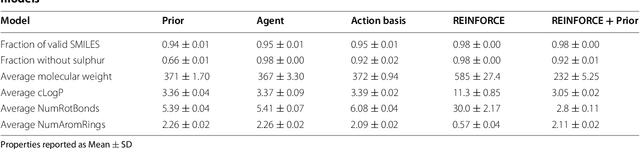
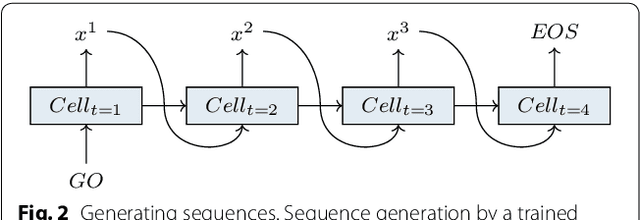

Abstract:This work introduces a method to tune a sequence-based generative model for molecular de novo design that through augmented episodic likelihood can learn to generate structures with certain specified desirable properties. We demonstrate how this model can execute a range of tasks such as generating analogues to a query structure and generating compounds predicted to be active against a biological target. As a proof of principle, the model is first trained to generate molecules that do not contain sulphur. As a second example, the model is trained to generate analogues to the drug Celecoxib, a technique that could be used for scaffold hopping or library expansion starting from a single molecule. Finally, when tuning the model towards generating compounds predicted to be active against the dopamine receptor type 2, the model generates structures of which more than 95% are predicted to be active, including experimentally confirmed actives that have not been included in either the generative model nor the activity prediction model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge