N. Duane Loh
HSG-12M: A Large-Scale Spatial Multigraph Dataset
Jun 10, 2025Abstract:Existing graph benchmarks assume non-spatial, simple edges, collapsing physically distinct paths into a single link. We introduce HSG-12M, the first large-scale dataset of $\textbf{spatial multigraphs}-$graphs embedded in a metric space where multiple geometrically distinct trajectories between two nodes are retained as separate edges. HSG-12M contains 11.6 million static and 5.1 million dynamic $\textit{Hamiltonian spectral graphs}$ across 1401 characteristic-polynomial classes, derived from 177 TB of spectral potential data. Each graph encodes the full geometry of a 1-D crystal's energy spectrum on the complex plane, producing diverse, physics-grounded topologies that transcend conventional node-coordinate datasets. To enable future extensions, we release $\texttt{Poly2Graph}$: a high-performance, open-source pipeline that maps arbitrary 1-D crystal Hamiltonians to spectral graphs. Benchmarks with popular GNNs expose new challenges in learning from multi-edge geometry at scale. Beyond its practical utility, we show that spectral graphs serve as universal topological fingerprints of polynomials, vectors, and matrices, forging a new algebra-to-graph link. HSG-12M lays the groundwork for geometry-aware graph learning and new opportunities of data-driven scientific discovery in condensed matter physics and beyond.
Single-shot pop-out 3D metrology of thin specimens with TEM
Sep 07, 2022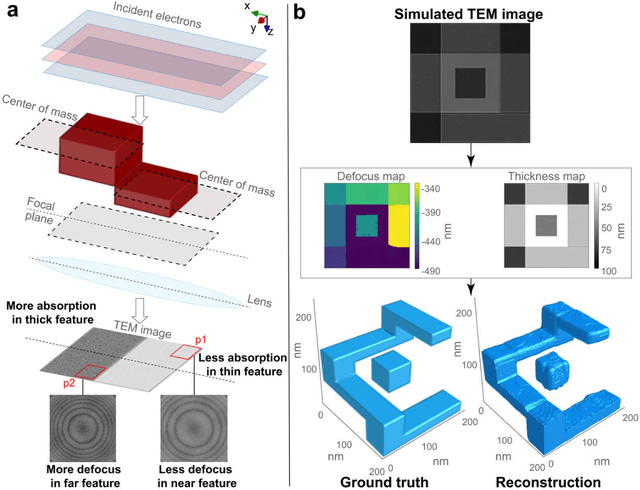
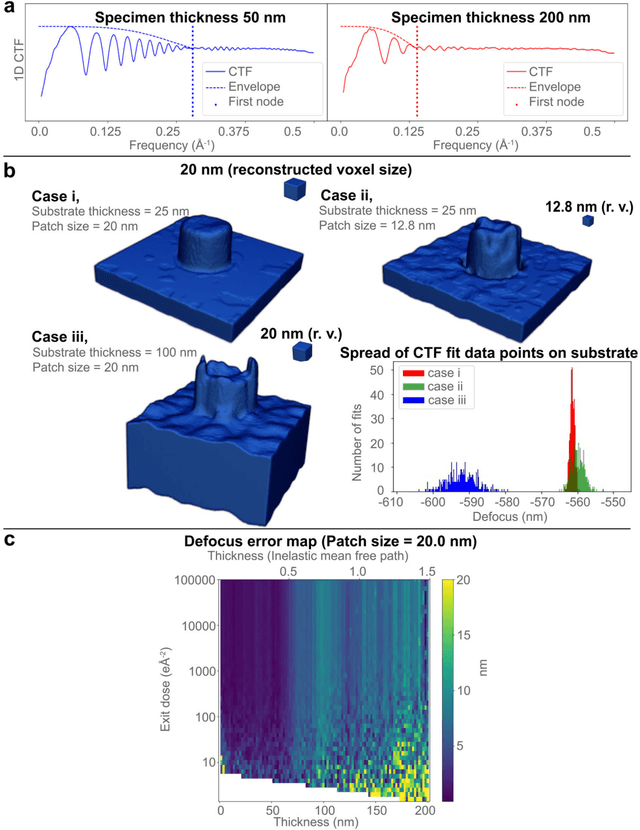
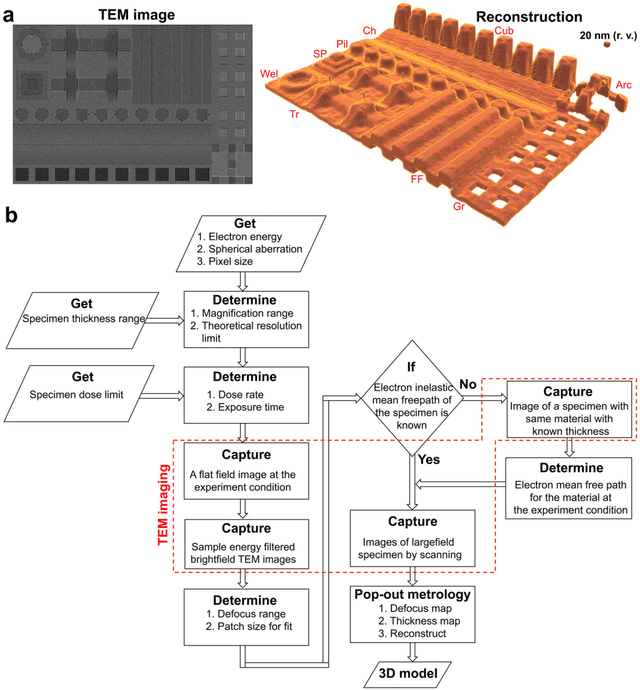
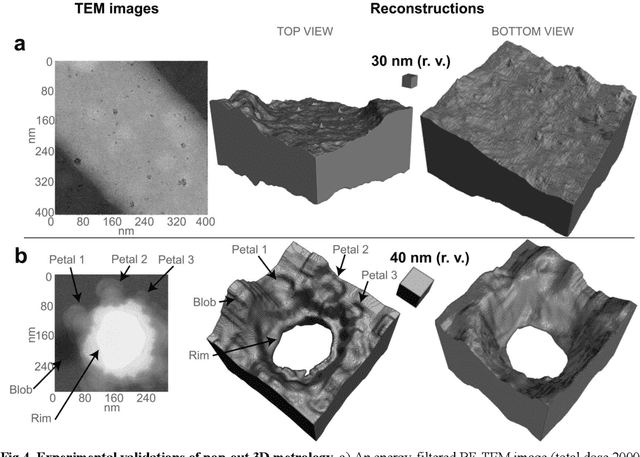
Abstract:Three-dimensional (3D) imaging of thin, extended specimens at nanometer resolution is critical for applications in biology, materials science, advanced synthesis, and manufacturing. Many 3D imaging techniques are limited to surface features, or available only for selective cross-sections, or require a tilt series of a local region, hence making them unsuitable for rapid, non-sacrificial screening of extended objects, or investigating fast dynamics. Here we describe a coherent imaging technique that recovers the 3D volume of a thin specimen with only a single, non-tomographic, energy-filtered, bright-field transmission electron microscopy (TEM) image. This technique does not require physically fracturing or sectioning thin specimens, only needs a single brief exposures to electron doses of ~100 e {\AA}-2, and can be readily calibrated for many existing TEMs; thus it can be widely deployed for rapid 3D metrology that complements existing forms of metrology.
Unsupervised learning approaches to characterize heterogeneous samples using X-ray single particle imaging
Sep 13, 2021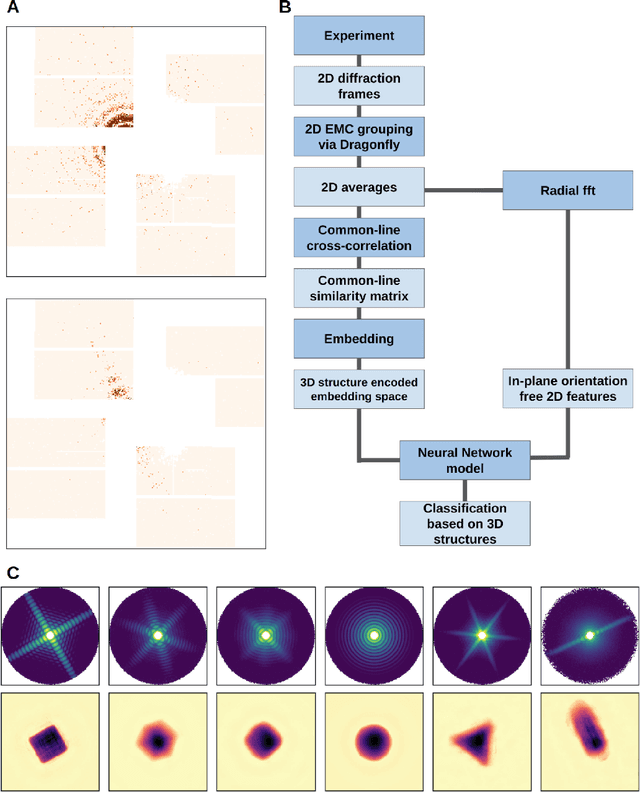
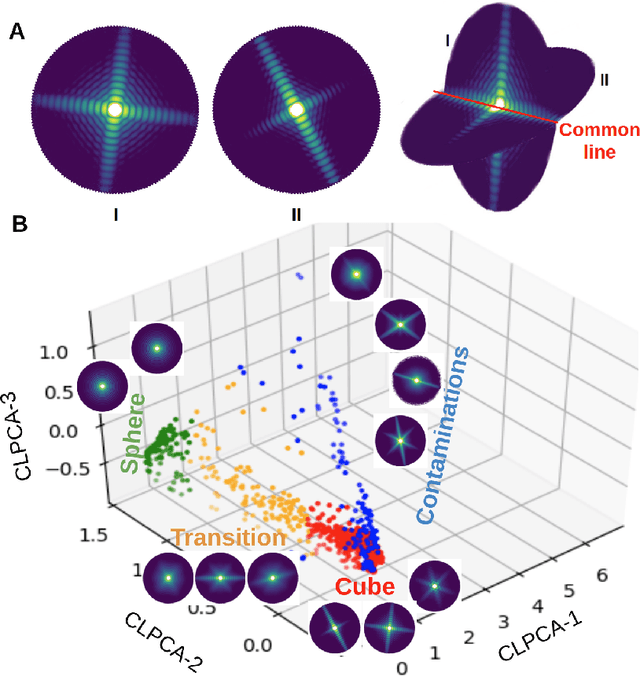
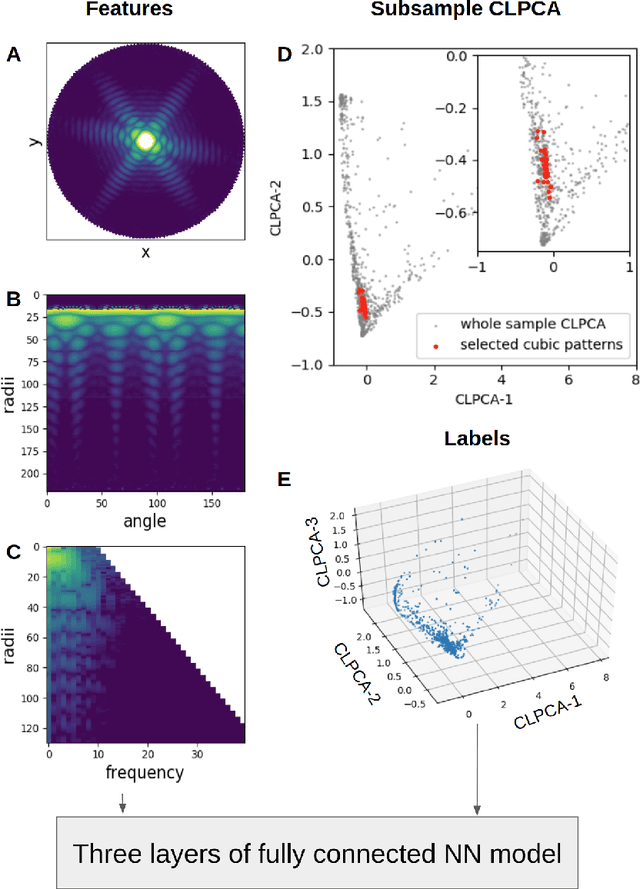
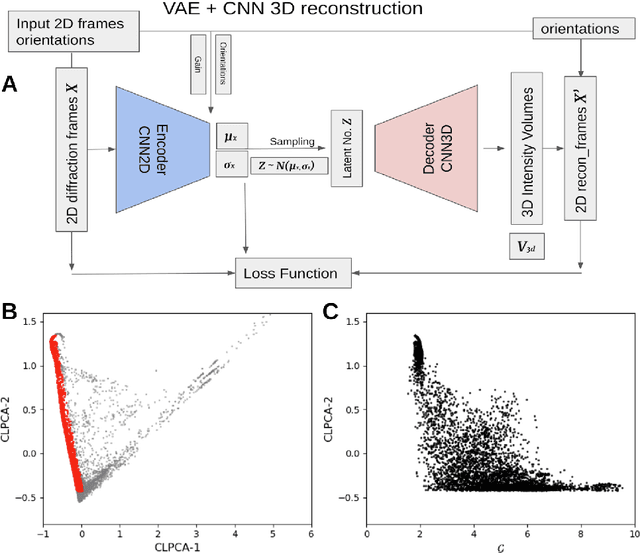
Abstract:One of the outstanding analytical problems in X-ray single particle imaging (SPI) is the classification of structural heterogeneity, which is especially difficult given the low signal-to-noise ratios of individual patterns and that even identical objects can yield patterns that vary greatly when orientation is taken into consideration. We propose two methods which explicitly account for this orientation-induced variation and can robustly determine the structural landscape of a sample ensemble. The first, termed common-line principal component analysis (PCA) provides a rough classification which is essentially parameter-free and can be run automatically on any SPI dataset. The second method, utilizing variation auto-encoders (VAEs) can generate 3D structures of the objects at any point in the structural landscape. We implement both these methods in combination with the noise-tolerant expand-maximize-compress (EMC) algorithm and demonstrate its utility by applying it to an experimental dataset from gold nanoparticles with only a few thousand photons per pattern and recover both discrete structural classes as well as continuous deformations. These developments diverge from previous approaches of extracting reproducible subsets of patterns from a dataset and open up the possibility to move beyond studying homogeneous sample sets and study open questions on topics such as nanocrystal growth and dynamics as well as phase transitions which have not been externally triggered.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge