Mohd Adnan
Residual GRU+MHSA: A Lightweight Hybrid Recurrent Attention Model for Cardiovascular Disease Detection
Dec 16, 2025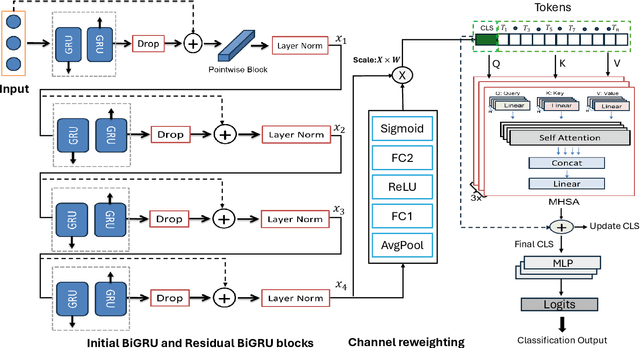
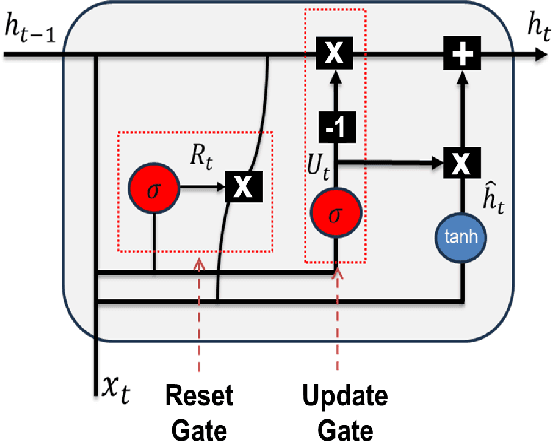
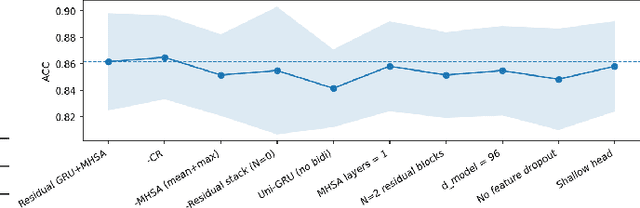
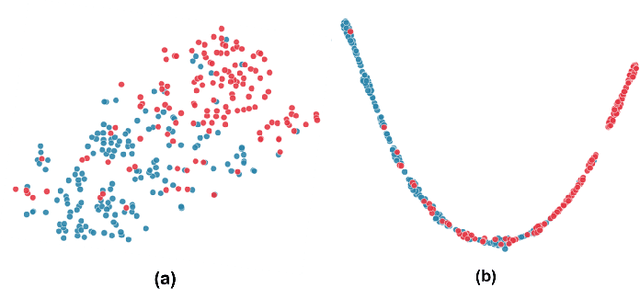
Abstract:Cardiovascular disease (CVD) remains the leading cause of mortality worldwide, underscoring the need for reliable and efficient predictive tools that support early intervention. Traditional diagnostic approaches rely on handcrafted features and clinician expertise, while machine learning methods improve reproducibility but often struggle to generalize across noisy and heterogeneous clinical data. In this work, we propose Residual GRU with Multi-Head Self-Attention, a compact deep learning architecture designed for tabular clinical records. The model integrates residual bidirectional gated recurrent units for sequential modeling of feature columns, a channel reweighting block, and multi-head self-attention pooling with a learnable classification token to capture global context. We evaluate the model on the UCI Heart Disease dataset using 5-fold stratified cross-validation and compare it against classical methods such as Logistic Regression, Random Forest, and Support Vector Machines, as well as modern deep learning baselines including DeepMLP, convolutional networks, recurrent networks, and Transformers. The proposed model achieves an accuracy of 0.861, macro-F1 of 0.860, ROC-AUC of 0.908, and PR-AUC of 0.904, outperforming all baselines. Ablation studies confirm the individual contributions of residual recurrence, channel gating, and attention pooling. t-SNE visualizations further indicate that the learned embeddings exhibit clearer separation between disease and non-disease classes compared to raw features. These results demonstrate that lightweight hybrid recurrent and attention-based architectures provide a strong balance between accuracy and efficiency for clinical risk prediction, supporting deployment in resource-constrained healthcare settings.
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021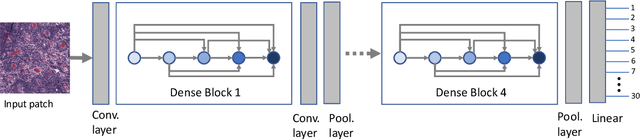
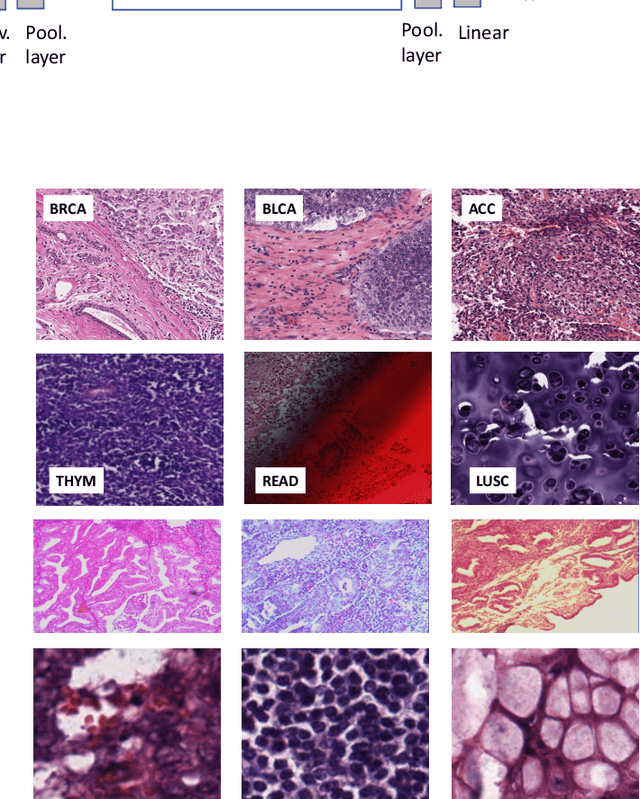
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge