Amir Safarpoor
Learning to Predict RNA Sequence Expressions from Whole Slide Images with Applications for Search and Classification
Mar 26, 2022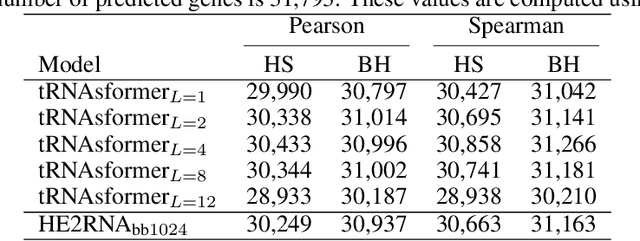
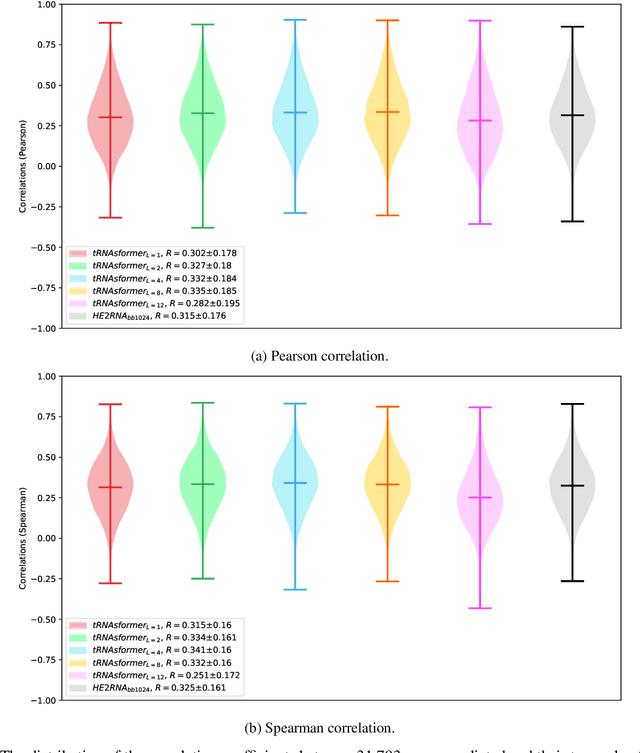
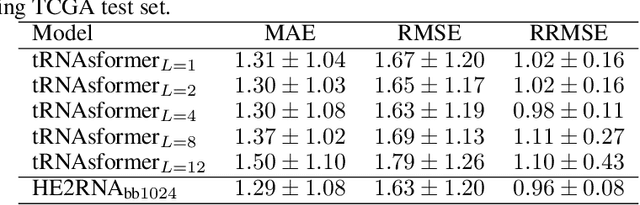
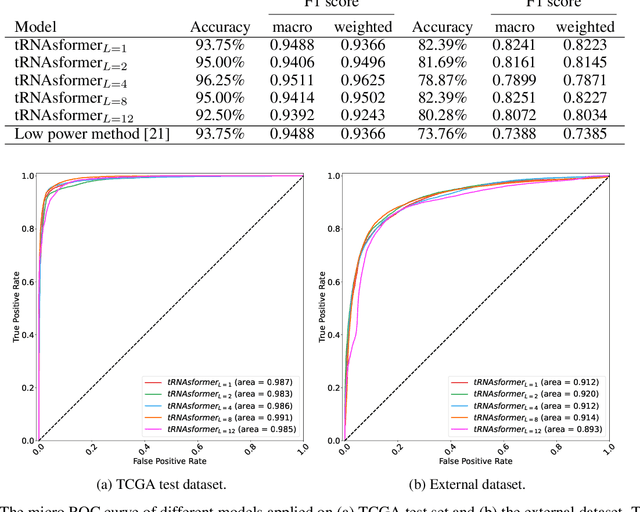
Abstract:Deep learning methods are widely applied in digital pathology to address clinical challenges such as prognosis and diagnosis. As one of the most recent applications, deep models have also been used to extract molecular features from whole slide images. Although molecular tests carry rich information, they are often expensive, time-consuming, and require additional tissue to sample. In this paper, we propose tRNAsfomer, an attention-based topology that can learn both to predict the bulk RNA-seq from an image and represent the whole slide image of a glass slide simultaneously. The tRNAsfomer uses multiple instance learning to solve a weakly supervised problem while the pixel-level annotation is not available for an image. We conducted several experiments and achieved better performance and faster convergence in comparison to the state-of-the-art algorithms. The proposed tRNAsfomer can assist as a computational pathology tool to facilitate a new generation of search and classification methods by combining the tissue morphology and the molecular fingerprint of the biopsy samples.
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021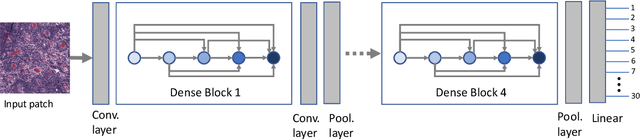
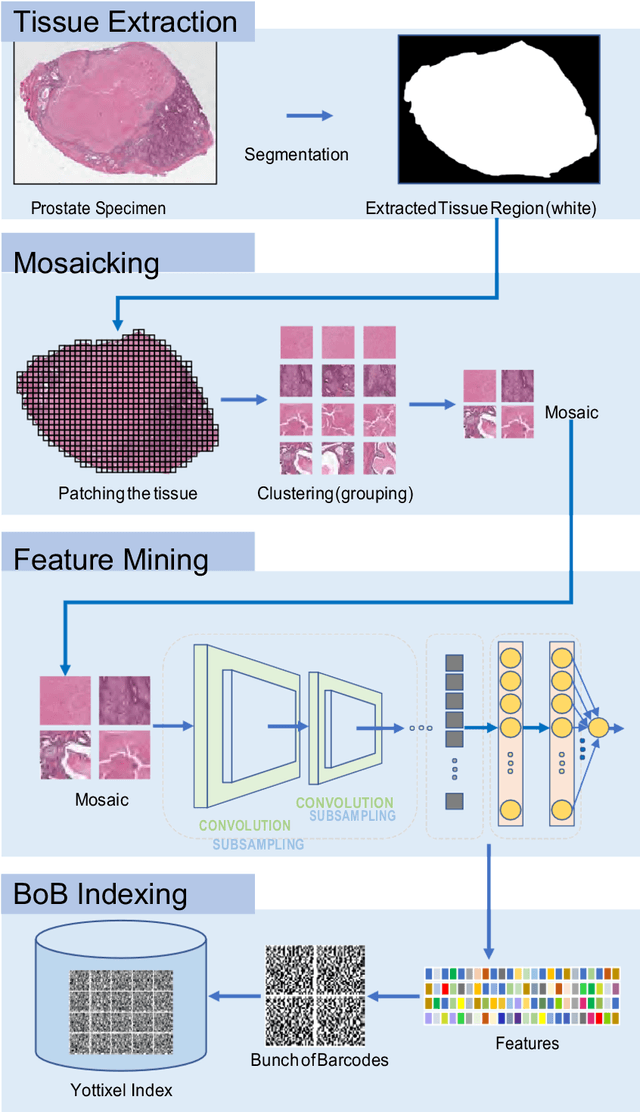
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
Offline versus Online Triplet Mining based on Extreme Distances of Histopathology Patches
Jul 04, 2020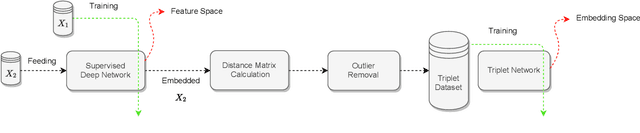

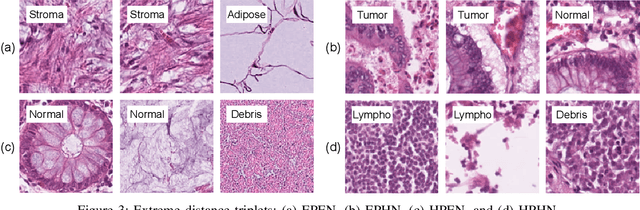
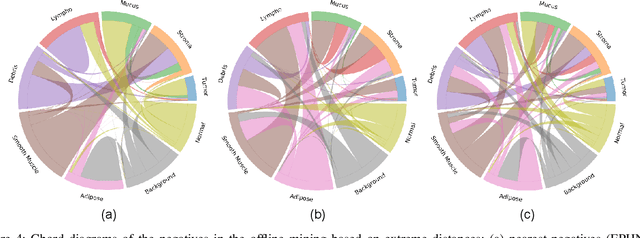
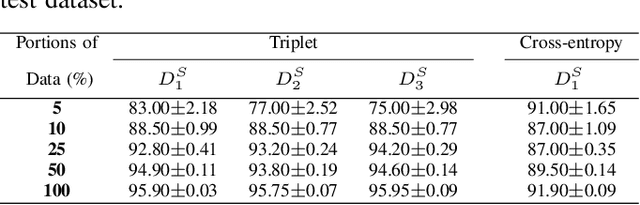
Abstract:We analyze the effect of offline and online triplet mining for colorectal cancer (CRC) histopathology dataset containing 100,000 patches. We consider the extreme, i.e., farthest and nearest patches with respect to a given anchor, both in online and offline mining. While many works focus solely on how to select the triplets online (batch-wise), we also study the effect of extreme distances and neighbor patches before training in an offline fashion. We analyze the impacts of extreme cases for offline versus online mining, including easy positive, batch semi-hard, and batch hard triplet mining as well as the neighborhood component analysis loss, its proxy version, and distance weighted sampling. We also investigate online approaches based on extreme distance and comprehensively compare the performance of offline and online mining based on the data patterns and explain offline mining as a tractable generalization of the online mining with large mini-batch size. As well, we discuss the relations of different colorectal tissue types in terms of extreme distances. We found that offline mining can generate a better statistical representation of the population by working on the whole dataset.
Supervision and Source Domain Impact on Representation Learning: A Histopathology Case Study
May 10, 2020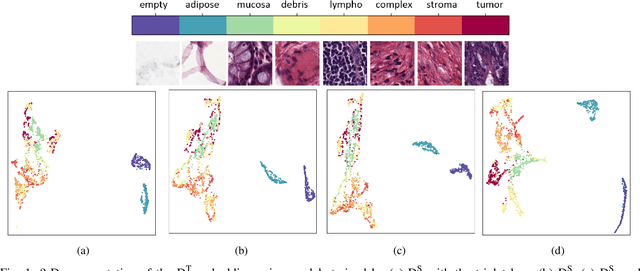
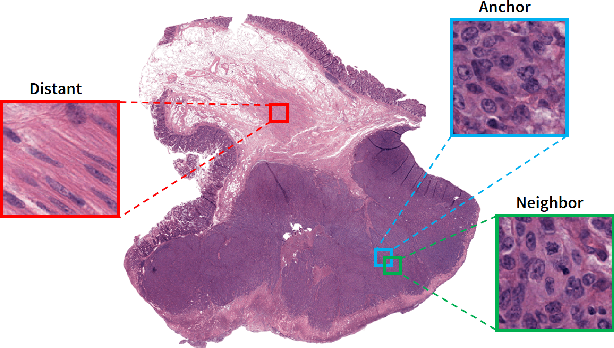

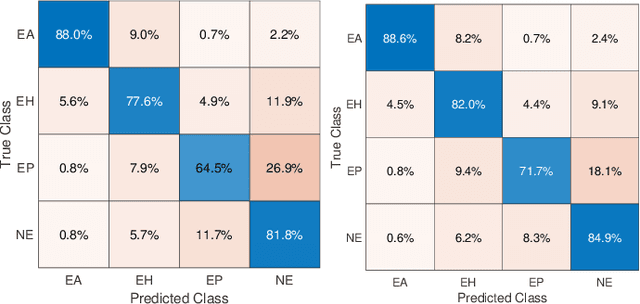
Abstract:As many algorithms depend on a suitable representation of data, learning unique features is considered a crucial task. Although supervised techniques using deep neural networks have boosted the performance of representation learning, the need for a large set of labeled data limits the application of such methods. As an example, high-quality delineations of regions of interest in the field of pathology is a tedious and time-consuming task due to the large image dimensions. In this work, we explored the performance of a deep neural network and triplet loss in the area of representation learning. We investigated the notion of similarity and dissimilarity in pathology whole-slide images and compared different setups from unsupervised and semi-supervised to supervised learning in our experiments. Additionally, different approaches were tested, applying few-shot learning on two publicly available pathology image datasets. We achieved high accuracy and generalization when the learned representations were applied to two different pathology datasets.
Pan-Cancer Diagnostic Consensus Through Searching Archival Histopathology Images Using Artificial Intelligence
Nov 20, 2019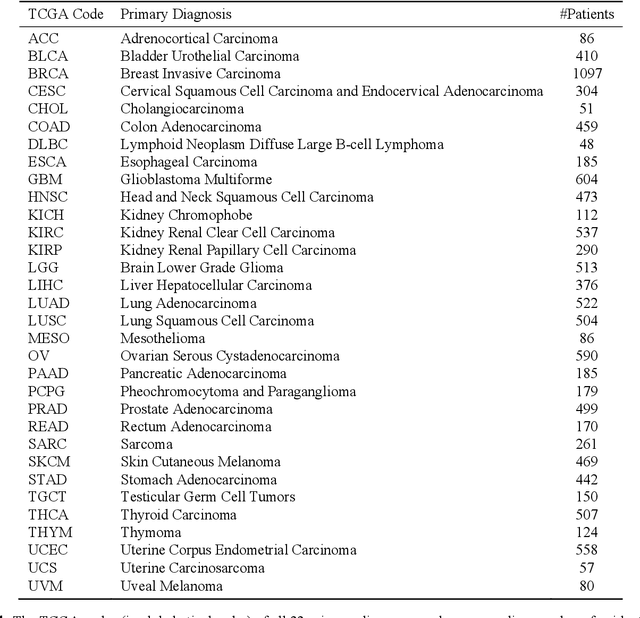
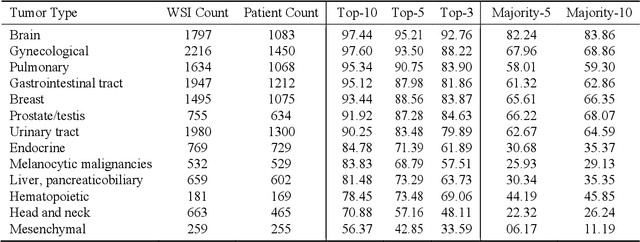
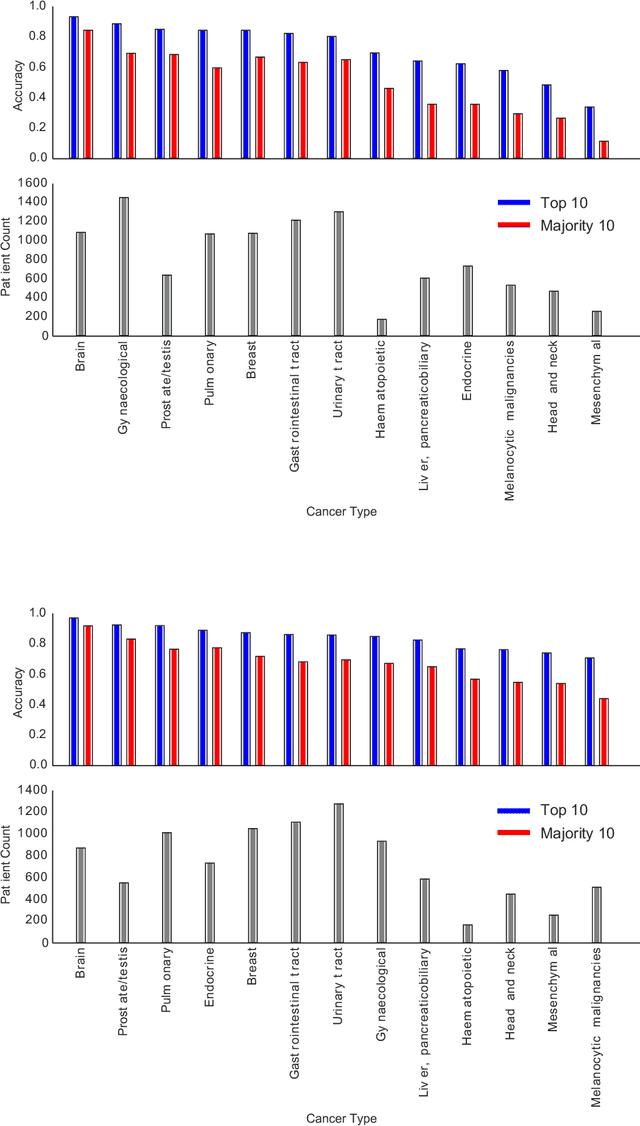
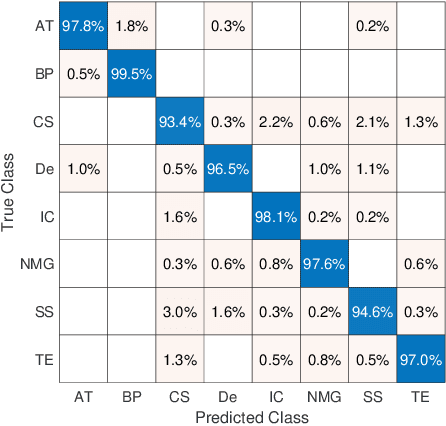
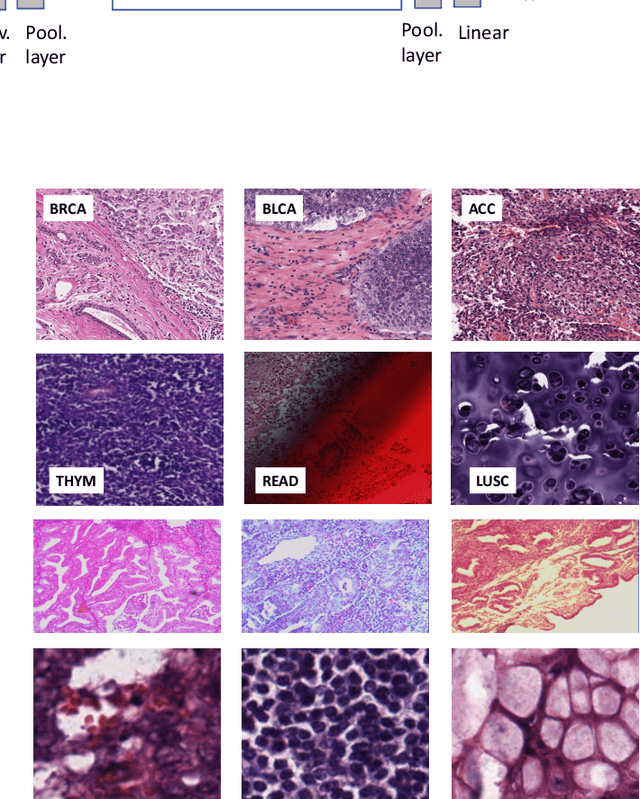
Abstract:The emergence of digital pathology has opened new horizons for histopathology and cytology. Artificial-intelligence algorithms are able to operate on digitized slides to assist pathologists with diagnostic tasks. Whereas machine learning involving classification and segmentation methods have obvious benefits for image analysis in pathology, image search represents a fundamental shift in computational pathology. Matching the pathology of new patients with already diagnosed and curated cases offers pathologist a novel approach to improve diagnostic accuracy through visual inspection of similar cases and computational majority vote for consensus building. In this study, we report the results from searching the largest public repository (The Cancer Genome Atlas [TCGA] program by National Cancer Institute, USA) of whole slide images from almost 11,000 patients depicting different types of malignancies. For the first time, we successfully indexed and searched almost 30,000 high-resolution digitized slides constituting 16 terabytes of data comprised of 20 million 1000x1000 pixels image patches. The TCGA image database covers 25 anatomic sites and contains 32 cancer subtypes. High-performance storage and GPU power were employed for experimentation. The results were assessed with conservative "majority voting" to build consensus for subtype diagnosis through vertical search and demonstrated high accuracy values for both frozen sections slides (e.g., bladder urothelial carcinoma 93%, kidney renal clear cell carcinoma 97%, and ovarian serous cystadenocarcinoma 99%) and permanent histopathology slides (e.g., prostate adenocarcinoma 98%, skin cutaneous melanoma 99%, and thymoma 100%). The key finding of this validation study was that computational consensus appears to be possible for rendering diagnoses if a sufficiently large number of searchable cases are available for each cancer subtype.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge