Minh Doan
SPAR: Session-based Pipeline for Adaptive Retrieval on Legacy File Systems
Dec 15, 2025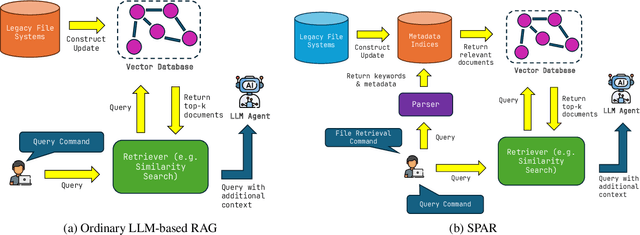

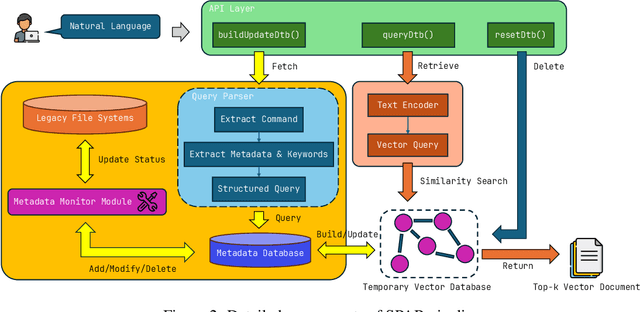
Abstract:The ability to extract value from historical data is essential for enterprise decision-making. However, much of this information remains inaccessible within large legacy file systems that lack structured organization and semantic indexing, making retrieval and analysis inefficient and error-prone. We introduce SPAR (Session-based Pipeline for Adaptive Retrieval), a conceptual framework that integrates Large Language Models (LLMs) into a Retrieval-Augmented Generation (RAG) architecture specifically designed for legacy enterprise environments. Unlike conventional RAG pipelines, which require costly construction and maintenance of full-scale vector databases that mirror the entire file system, SPAR employs a lightweight two-stage process: a semantic Metadata Index is first created, after which session-specific vector databases are dynamically generated on demand. This design reduces computational overhead while improving transparency, controllability, and relevance in retrieval. We provide a theoretical complexity analysis comparing SPAR with standard LLM-based RAG pipelines, demonstrating its computational advantages. To validate the framework, we apply SPAR to a synthesized enterprise-scale file system containing a large corpus of biomedical literature, showing improvements in both retrieval effectiveness and downstream model accuracy. Finally, we discuss design trade-offs and outline open challenges for deploying SPAR across diverse enterprise settings.
Can virtual staining for high-throughput screening generalize?
Jul 09, 2024



Abstract:The large volume and variety of imaging data from high-throughput screening (HTS) in the pharmaceutical industry present an excellent resource for training virtual staining models. However, the potential of models trained under one set of experimental conditions to generalize to other conditions remains underexplored. This study systematically investigates whether data from three cell types (lung, ovarian, and breast) and two phenotypes (toxic and non-toxic conditions) commonly found in HTS can effectively train virtual staining models to generalize across three typical HTS distribution shifts: unseen phenotypes, unseen cell types, and the combination of both. Utilizing a dataset of 772,416 paired bright-field, cytoplasm, nuclei, and DNA-damage stain images, we evaluate the generalization capabilities of models across pixel-based, instance-wise, and biological-feature-based levels. Our findings indicate that training virtual nuclei and cytoplasm models on non-toxic condition samples not only generalizes to toxic condition samples but leads to improved performance across all evaluation levels compared to training on toxic condition samples. Generalization to unseen cell types shows variability depending on the cell type; models trained on ovarian or lung cell samples often perform well under other conditions, while those trained on breast cell samples consistently show poor generalization. Generalization to unseen cell types and phenotypes shows good generalization across all levels of evaluation compared to addressing unseen cell types alone. This study represents the first large-scale, data-centric analysis of the generalization capability of virtual staining models trained on diverse HTS datasets, providing valuable strategies for experimental training data generation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge