Mingyuan Meng
Merging-Diverging Hybrid Transformer Networks for Survival Prediction in Head and Neck Cancer
Jul 07, 2023Abstract:Survival prediction is crucial for cancer patients as it provides early prognostic information for treatment planning. Recently, deep survival models based on deep learning and medical images have shown promising performance for survival prediction. However, existing deep survival models are not well developed in utilizing multi-modality images (e.g., PET-CT) and in extracting region-specific information (e.g., the prognostic information in Primary Tumor (PT) and Metastatic Lymph Node (MLN) regions). In view of this, we propose a merging-diverging learning framework for survival prediction from multi-modality images. This framework has a merging encoder to fuse multi-modality information and a diverging decoder to extract region-specific information. In the merging encoder, we propose a Hybrid Parallel Cross-Attention (HPCA) block to effectively fuse multi-modality features via parallel convolutional layers and cross-attention transformers. In the diverging decoder, we propose a Region-specific Attention Gate (RAG) block to screen out the features related to lesion regions. Our framework is demonstrated on survival prediction from PET-CT images in Head and Neck (H&N) cancer, by designing an X-shape merging-diverging hybrid transformer network (named XSurv). Our XSurv combines the complementary information in PET and CT images and extracts the region-specific prognostic information in PT and MLN regions. Extensive experiments on the public dataset of HEad and neCK TumOR segmentation and outcome prediction challenge (HECKTOR 2022) demonstrate that our XSurv outperforms state-of-the-art survival prediction methods.
Non-iterative Coarse-to-fine Transformer Networks for Joint Affine and Deformable Image Registration
Jul 07, 2023Abstract:Image registration is a fundamental requirement for medical image analysis. Deep registration methods based on deep learning have been widely recognized for their capabilities to perform fast end-to-end registration. Many deep registration methods achieved state-of-the-art performance by performing coarse-to-fine registration, where multiple registration steps were iterated with cascaded networks. Recently, Non-Iterative Coarse-to-finE (NICE) registration methods have been proposed to perform coarse-to-fine registration in a single network and showed advantages in both registration accuracy and runtime. However, existing NICE registration methods mainly focus on deformable registration, while affine registration, a common prerequisite, is still reliant on time-consuming traditional optimization-based methods or extra affine registration networks. In addition, existing NICE registration methods are limited by the intrinsic locality of convolution operations. Transformers may address this limitation for their capabilities to capture long-range dependency, but the benefits of using transformers for NICE registration have not been explored. In this study, we propose a Non-Iterative Coarse-to-finE Transformer network (NICE-Trans) for image registration. Our NICE-Trans is the first deep registration method that (i) performs joint affine and deformable coarse-to-fine registration within a single network, and (ii) embeds transformers into a NICE registration framework to model long-range relevance between images. Extensive experiments with seven public datasets show that our NICE-Trans outperforms state-of-the-art registration methods on both registration accuracy and runtime.
DeepMSS: Deep Multi-Modality Segmentation-to-Survival Learning for Survival Outcome Prediction from PET/CT Images
May 17, 2023Abstract:Survival prediction is a major concern for cancer management. Deep survival models based on deep learning have been widely adopted to perform end-to-end survival prediction from medical images. Recent deep survival models achieved promising performance by jointly performing tumor segmentation with survival prediction, where the models were guided to extract tumor-related information through Multi-Task Learning (MTL). However, existing deep survival models have difficulties in exploring out-of-tumor prognostic information (e.g., local lymph node metastasis and adjacent tissue invasions). In addition, existing deep survival models are underdeveloped in utilizing multi-modality images. Empirically-designed strategies were commonly adopted to fuse multi-modality information via fixed pre-designed networks. In this study, we propose a Deep Multi-modality Segmentation-to-Survival model (DeepMSS) for survival prediction from PET/CT images. Instead of adopting MTL, we propose a novel Segmentation-to-Survival Learning (SSL) strategy, where our DeepMSS is trained for tumor segmentation and survival prediction sequentially. This strategy enables the DeepMSS to initially focus on tumor regions and gradually expand its focus to include other prognosis-related regions. We also propose a data-driven strategy to fuse multi-modality image information, which realizes automatic optimization of fusion strategies based on training data during training and also improves the adaptability of DeepMSS to different training targets. Our DeepMSS is also capable of incorporating conventional radiomics features as an enhancement, where handcrafted features can be extracted from the DeepMSS-segmented tumor regions and cooperatively integrated into the DeepMSS's training and inference. Extensive experiments with two large clinical datasets show that our DeepMSS outperforms state-of-the-art survival prediction methods.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Brain Tumor Sequence Registration with Non-iterative Coarse-to-fine Networks and Dual Deep Supervision
Nov 15, 2022Abstract:In this study, we focus on brain tumor sequence registration between pre-operative and follow-up Magnetic Resonance Imaging (MRI) scans of brain glioma patients, in the context of Brain Tumor Sequence Registration challenge (BraTS-Reg 2022). Brain tumor registration is a fundamental requirement in brain image analysis for quantifying tumor changes. This is a challenging task due to large deformations and missing correspondences between pre-operative and follow-up scans. For this task, we adopt our recently proposed Non-Iterative Coarse-to-finE registration Networks (NICE-Net) - a deep learning-based method for coarse-to-fine registering images with large deformations. To overcome missing correspondences, we extend the NICE-Net by introducing dual deep supervision, where a deep self-supervised loss based on image similarity and a deep weakly-supervised loss based on manually annotated landmarks are deeply embedded into the NICE-Net. At the BraTS-Reg 2022, our method achieved a competitive result on the validation set (mean absolute error: 3.387) and placed 4th in the final testing phase (Score: 0.3544).
Radiomics-enhanced Deep Multi-task Learning for Outcome Prediction in Head and Neck Cancer
Nov 10, 2022Abstract:Outcome prediction is crucial for head and neck cancer patients as it can provide prognostic information for early treatment planning. Radiomics methods have been widely used for outcome prediction from medical images. However, these methods are limited by their reliance on intractable manual segmentation of tumor regions. Recently, deep learning methods have been proposed to perform end-to-end outcome prediction so as to remove the reliance on manual segmentation. Unfortunately, without segmentation masks, these methods will take the whole image as input, such that makes them difficult to focus on tumor regions and potentially unable to fully leverage the prognostic information within the tumor regions. In this study, we propose a radiomics-enhanced deep multi-task framework for outcome prediction from PET/CT images, in the context of HEad and neCK TumOR segmentation and outcome prediction challenge (HECKTOR 2022). In our framework, our novelty is to incorporate radiomics as an enhancement to our recently proposed Deep Multi-task Survival model (DeepMTS). The DeepMTS jointly learns to predict the survival risk scores of patients and the segmentation masks of tumor regions. Radiomics features are extracted from the predicted tumor regions and combined with the predicted survival risk scores for final outcome prediction, through which the prognostic information in tumor regions can be further leveraged. Our method achieved a C-index of 0.681 on the testing set, placing the 2nd on the leaderboard with only 0.00068 lower in C-index than the 1st place.
Non-iterative Coarse-to-fine Registration based on Single-pass Deep Cumulative Learning
Jun 30, 2022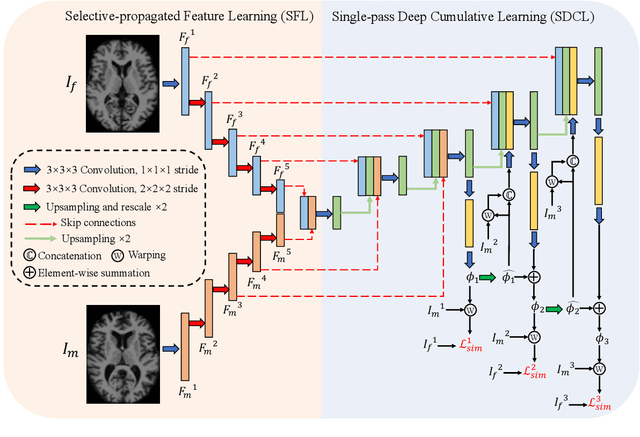
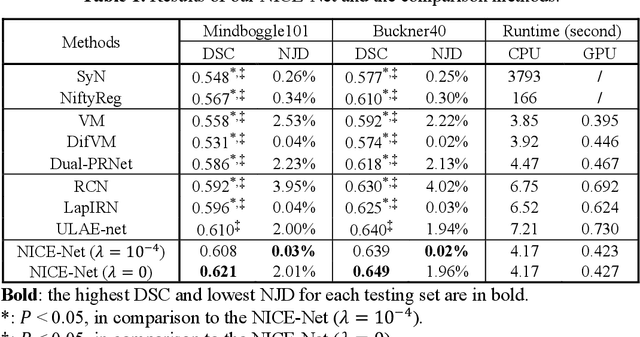
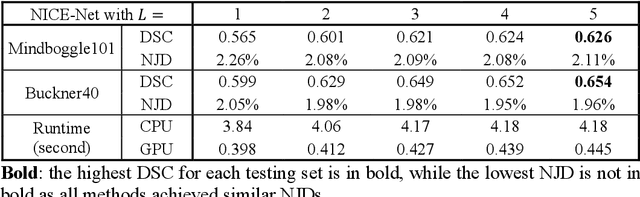
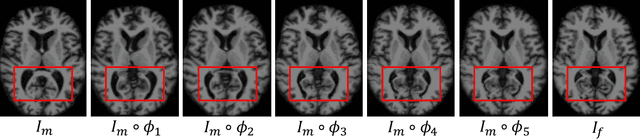
Abstract:Deformable image registration is a crucial step in medical image analysis for finding a non-linear spatial transformation between a pair of fixed and moving images. Deep registration methods based on Convolutional Neural Networks (CNNs) have been widely used as they can perform image registration in a fast and end-to-end manner. However, these methods usually have limited performance for image pairs with large deformations. Recently, iterative deep registration methods have been used to alleviate this limitation, where the transformations are iteratively learned in a coarse-to-fine manner. However, iterative methods inevitably prolong the registration runtime, and tend to learn separate image features for each iteration, which hinders the features from being leveraged to facilitate the registration at later iterations. In this study, we propose a Non-Iterative Coarse-to-finE registration Network (NICE-Net) for deformable image registration. In the NICE-Net, we propose: (i) a Single-pass Deep Cumulative Learning (SDCL) decoder that can cumulatively learn coarse-to-fine transformations within a single pass (iteration) of the network, and (ii) a Selectively-propagated Feature Learning (SFL) encoder that can learn common image features for the whole coarse-to-fine registration process and selectively propagate the features as needed. Extensive experiments on six public datasets of 3D brain Magnetic Resonance Imaging (MRI) show that our proposed NICE-Net can outperform state-of-the-art iterative deep registration methods while only requiring similar runtime to non-iterative methods.
DeepMTS: Deep Multi-task Learning for Survival Prediction in Patients with Advanced Nasopharyngeal Carcinoma using Pretreatment PET/CT
Sep 16, 2021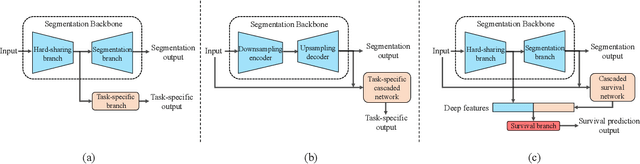
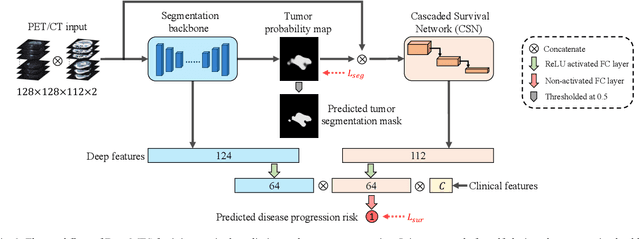
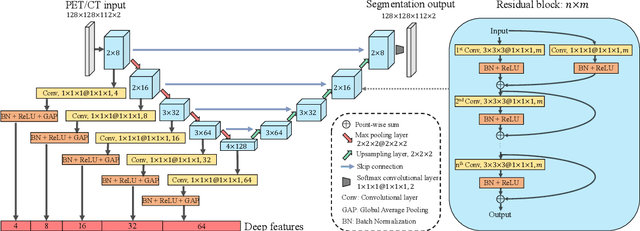
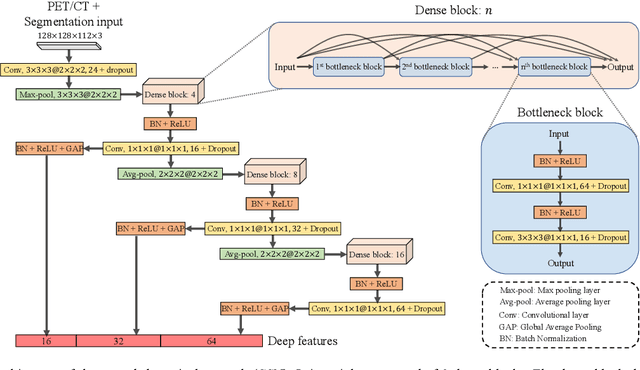
Abstract:Nasopharyngeal Carcinoma (NPC) is a worldwide malignant epithelial cancer. Survival prediction is a major concern for NPC patients, as it provides early prognostic information that is needed to guide treatments. Recently, deep learning, which leverages Deep Neural Networks (DNNs) to learn deep representations of image patterns, has been introduced to the survival prediction in various cancers including NPC. It has been reported that image-derived end-to-end deep survival models have the potential to outperform clinical prognostic indicators and traditional radiomics-based survival models in prognostic performance. However, deep survival models, especially 3D models, require large image training data to avoid overfitting. Unfortunately, medical image data is usually scarce, especially for Positron Emission Tomography/Computed Tomography (PET/CT) due to the high cost of PET/CT scanning. Compared to Magnetic Resonance Imaging (MRI) or Computed Tomography (CT) providing only anatomical information of tumors, PET/CT that provides both anatomical (from CT) and metabolic (from PET) information is promising to achieve more accurate survival prediction. However, we have not identified any 3D end-to-end deep survival model that applies to small PET/CT data of NPC patients. In this study, we introduced the concept of multi-task leaning into deep survival models to address the overfitting problem resulted from small data. Tumor segmentation was incorporated as an auxiliary task to enhance the model's efficiency of learning from scarce PET/CT data. Based on this idea, we proposed a 3D end-to-end Deep Multi-Task Survival model (DeepMTS) for joint survival prediction and tumor segmentation. Our DeepMTS can jointly learn survival prediction and tumor segmentation using PET/CT data of only 170 patients with advanced NPC.
Prediction of 5-year Progression-Free Survival in Advanced Nasopharyngeal Carcinoma with Pretreatment PET/CT using Multi-Modality Deep Learning-based Radiomics
Mar 09, 2021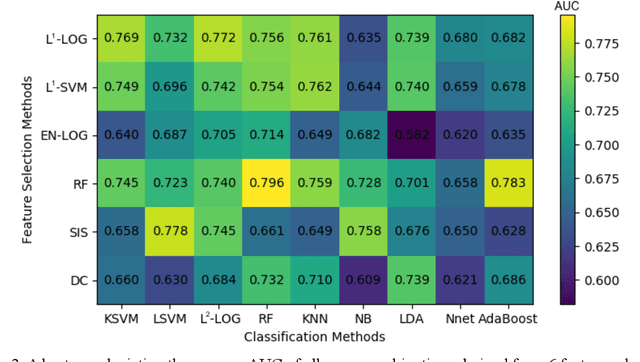
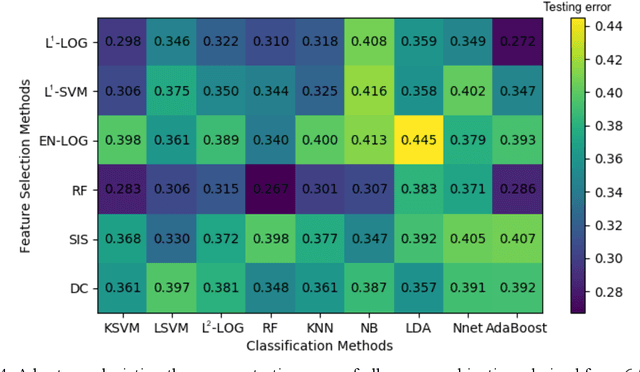
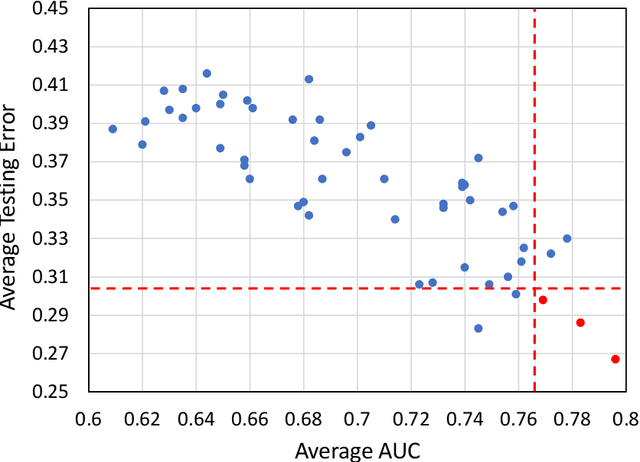
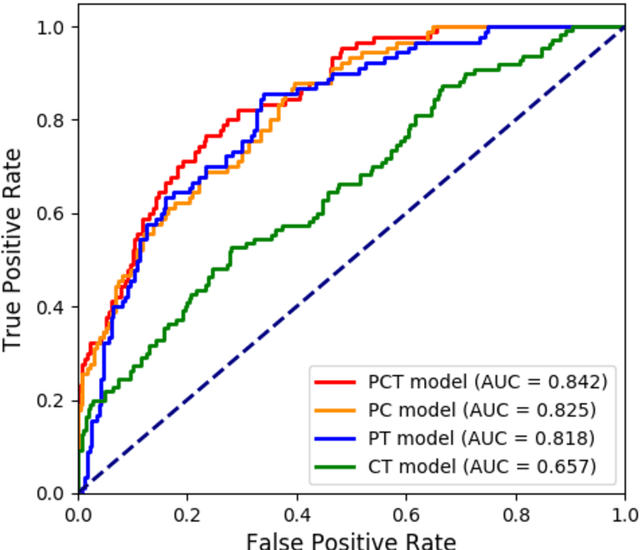
Abstract:Deep Learning-based Radiomics (DLR) has achieved great success on medical image analysis. In this study, we aim to explore the capability of DLR for survival prediction in NPC. We developed an end-to-end multi-modality DLR model using pretreatment PET/CT images to predict 5-year Progression-Free Survival (PFS) in advanced NPC. A total of 170 patients with pathological confirmed advanced NPC (TNM stage III or IVa) were enrolled in this study. A 3D Convolutional Neural Network (CNN), with two branches to process PET and CT separately, was optimized to extract deep features from pretreatment multi-modality PET/CT images and use the derived features to predict the probability of 5-year PFS. Optionally, TNM stage, as a high-level clinical feature, can be integrated into our DLR model to further improve prognostic performance. For a comparison between CR and DLR, 1456 handcrafted features were extracted, and three top CR methods were selected as benchmarks from 54 combinations of 6 feature selection methods and 9 classification methods. Compared to the three CR methods, our multi-modality DLR models using both PET and CT, with or without TNM stage (named PCT or PC model), resulted in the highest prognostic performance. Furthermore, the multi-modality PCT model outperformed single-modality DLR models using only PET and TNM stage (PT model) or only CT and TNM stage (CT model). Our study identified potential radiomics-based prognostic model for survival prediction in advanced NPC, and suggests that DLR could serve as a tool for aiding in cancer management.
Enhancing Medical Image Registration via Appearance Adjustment Networks
Mar 09, 2021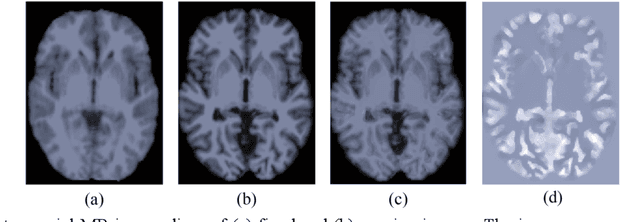
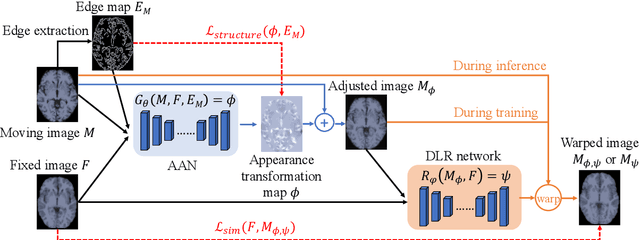
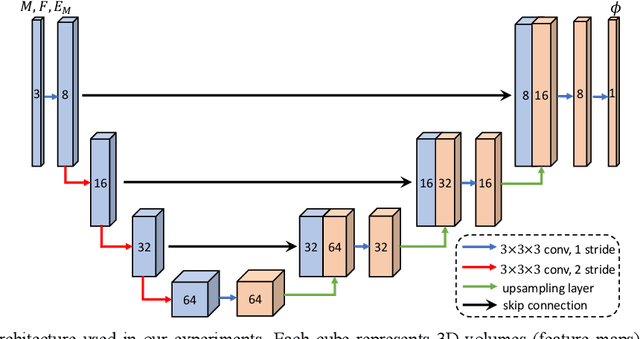
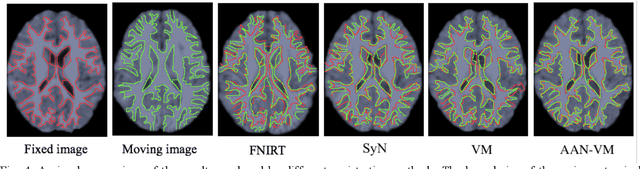
Abstract:Deformable image registration is fundamental for many medical image analyses. A key obstacle for accurate image registration is the variations in image appearance. Recently, deep learning-based registration methods (DLRs), using deep neural networks, have computational efficiency that is several orders of magnitude greater than traditional optimization-based registration methods (ORs). A major drawback, however, of DLRs is a disregard for the target-pair-specific optimization that is inherent in ORs and instead they rely on a globally optimized network that is trained with a set of training samples to achieve faster registration. Thus, DLRs inherently have degraded ability to adapt to appearance variations and perform poorly, compared to ORs, when image pairs (fixed/moving images) have large differences in appearance. Hence, we propose an Appearance Adjustment Network (AAN) where we leverage anatomy edges, through an anatomy-constrained loss function, to generate an anatomy-preserving appearance transformation. We designed the AAN so that it can be readily inserted into a wide range of DLRs, to reduce the appearance differences between the fixed and moving images. Our AAN and DLR's network can be trained cooperatively in an unsupervised and end-to-end manner. We evaluated our AAN with two widely used DLRs - Voxelmorph (VM) and FAst IMage registration (FAIM) - on three public 3D brain magnetic resonance (MR) image datasets - IBSR18, Mindboggle101, and LPBA40. The results show that DLRs, using the AAN, improved performance and achieved higher results than state-of-the-art ORs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge