Michael Mooney
Prospects for Theranostics in Neurosurgical Imaging: Empowering Confocal Laser Endomicroscopy Diagnostics via Deep Learning
Aug 18, 2018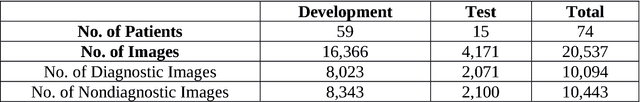
Abstract:Confocal laser endomicroscopy (CLE) is an advanced optical fluorescence imaging technology that has the potential to increase intraoperative precision, extend resection, and tailor surgery for malignant invasive brain tumors because of its subcellular dimension resolution. Despite its promising diagnostic potential, interpreting the gray tone fluorescence images can be difficult for untrained users. In this review, we provide a detailed description of bioinformatical analysis methodology of CLE images that begins to assist the neurosurgeon and pathologist to rapidly connect on-the-fly intraoperative imaging, pathology, and surgical observation into a conclusionary system within the concept of theranostics. We present an overview and discuss deep learning models for automatic detection of the diagnostic CLE images and discuss various training regimes and ensemble modeling effect on the power of deep learning predictive models. Two major approaches reviewed in this paper include the models that can automatically classify CLE images into diagnostic/nondiagnostic, glioma/nonglioma, tumor/injury/normal categories and models that can localize histological features on the CLE images using weakly supervised methods. We also briefly review advances in the deep learning approaches used for CLE image analysis in other organs. Significant advances in speed and precision of automated diagnostic frame selection would augment the diagnostic potential of CLE, improve operative workflow and integration into brain tumor surgery. Such technology and bioinformatics analytics lend themselves to improved precision, personalization, and theranostics in brain tumor treatment.
Convolutional Neural Networks: Ensemble Modeling, Fine-Tuning and Unsupervised Semantic Localization for Intraoperative CLE Images
Oct 23, 2017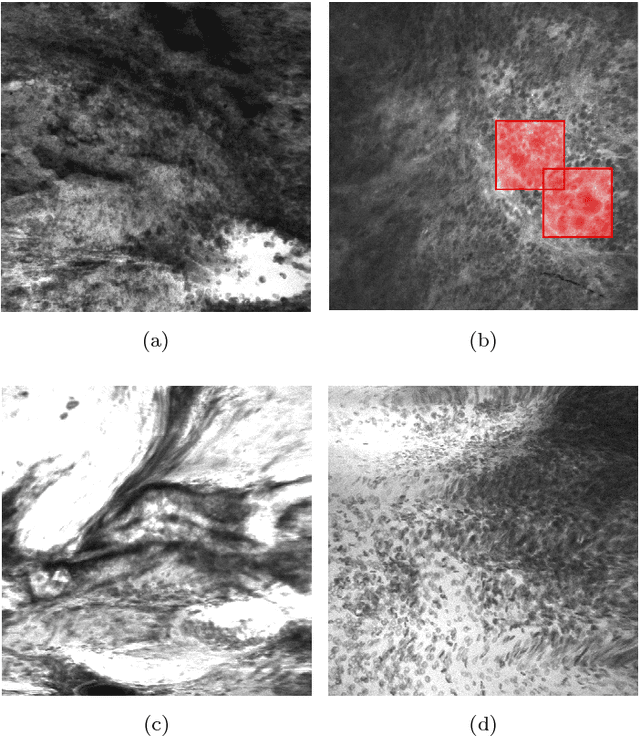
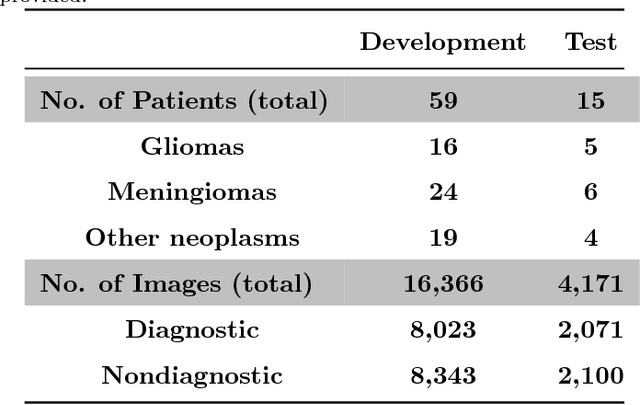
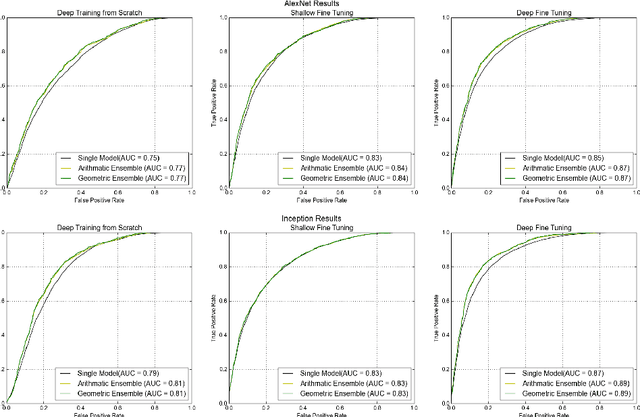
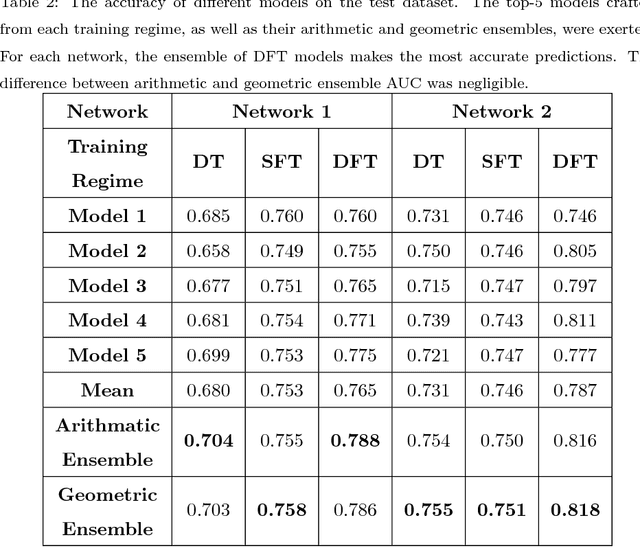
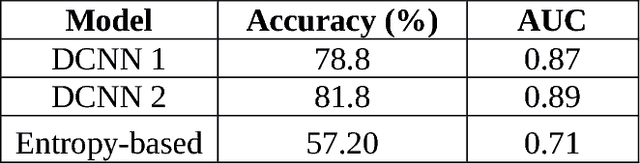
Abstract:Confocal laser endomicroscopy (CLE) is an advanced optical fluorescence technology undergoing assessment for applications in brain tumor surgery. Despite its promising potential, interpreting the unfamiliar gray tone images of fluorescent stains can be difficult. Many of the CLE images can be distorted by motion, extremely low or high fluorescence signal, or obscured by red blood cell accumulation, and these can be interpreted as nondiagnostic. However, just one neat CLE image might suffice for intraoperative diagnosis of the tumor. While manual examination of thousands of nondiagnostic images during surgery would be impractical, this creates an opportunity for a model to select diagnostic images for the pathologists or surgeon's review. In this study, we sought to develop a deep learning model to automatically detect the diagnostic images using a manually annotated dataset, and we employed a patient-based nested cross-validation approach to explore generalizability of the model. We explored various training regimes: deep training, shallow fine-tuning, and deep fine-tuning. Further, we investigated the effect of ensemble modeling by combining the top-5 single models crafted in the development phase. We localized histological features from diagnostic CLE images by visualization of shallow and deep neural activations. Our inter-rater experiment results confirmed that our ensemble of deeply fine-tuned models achieved higher agreement with the ground truth than the other observers. With the speed and precision of the proposed method (110 images/second; 85% on the gold standard test subset), it has potential to be integrated into the operative workflow in the brain tumor surgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge