Matteo Turilli
Scalable Runtime Architecture for Data-driven, Hybrid HPC and ML Workflow Applications
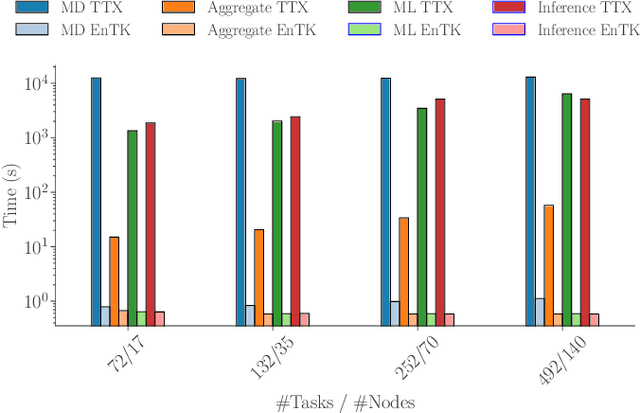
Mar 17, 2025Abstract:Hybrid workflows combining traditional HPC and novel ML methodologies are transforming scientific computing. This paper presents the architecture and implementation of a scalable runtime system that extends RADICAL-Pilot with service-based execution to support AI-out-HPC workflows. Our runtime system enables distributed ML capabilities, efficient resource management, and seamless HPC/ML coupling across local and remote platforms. Preliminary experimental results show that our approach manages concurrent execution of ML models across local and remote HPC/cloud resources with minimal architectural overheads. This lays the foundation for prototyping three representative data-driven workflow applications and executing them at scale on leadership-class HPC platforms.
AI-coupled HPC Workflows
Aug 24, 2022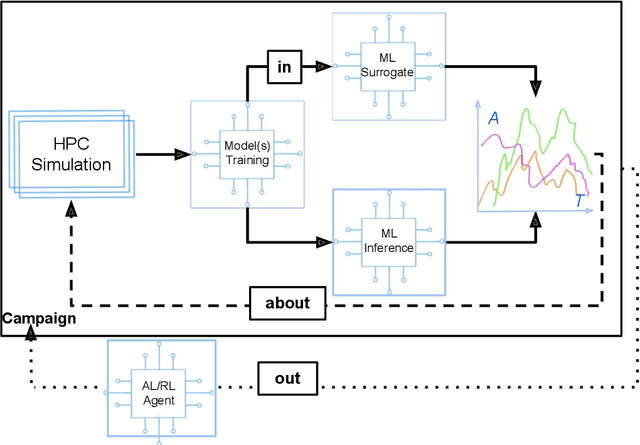
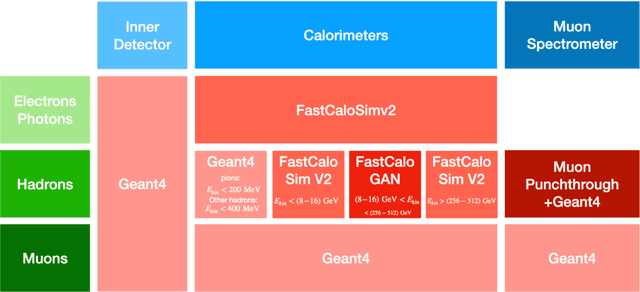
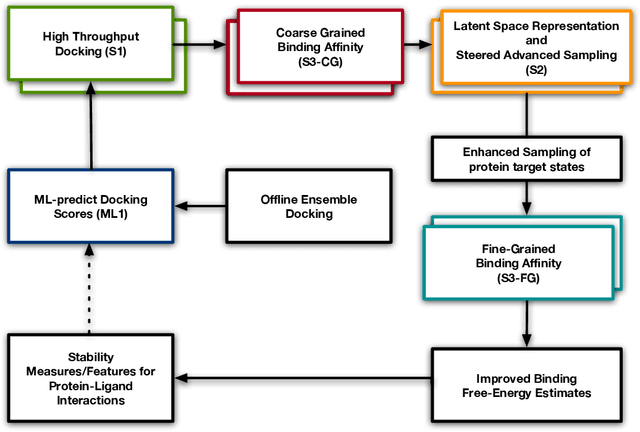
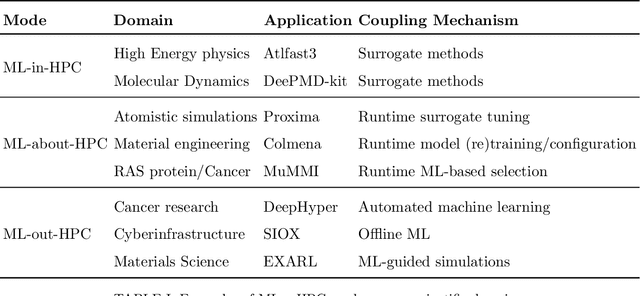
Abstract:Increasingly, scientific discovery requires sophisticated and scalable workflows. Workflows have become the ``new applications,'' wherein multi-scale computing campaigns comprise multiple and heterogeneous executable tasks. In particular, the introduction of AI/ML models into the traditional HPC workflows has been an enabler of highly accurate modeling, typically reducing computational needs compared to traditional methods. This chapter discusses various modes of integrating AI/ML models to HPC computations, resulting in diverse types of AI-coupled HPC workflows. The increasing need of coupling AI/ML and HPC across scientific domains is motivated, and then exemplified by a number of production-grade use cases for each mode. We additionally discuss the primary challenges of extreme-scale AI-coupled HPC campaigns -- task heterogeneity, adaptivity, performance -- and several framework and middleware solutions which aim to address them. While both HPC workflow and AI/ML computing paradigms are independently effective, we highlight how their integration, and ultimate convergence, is leading to significant improvements in scientific performance across a range of domains, ultimately resulting in scientific explorations otherwise unattainable.
Asynchronous Execution of Heterogeneous Tasks in AI-coupled HPC Workflows
Aug 23, 2022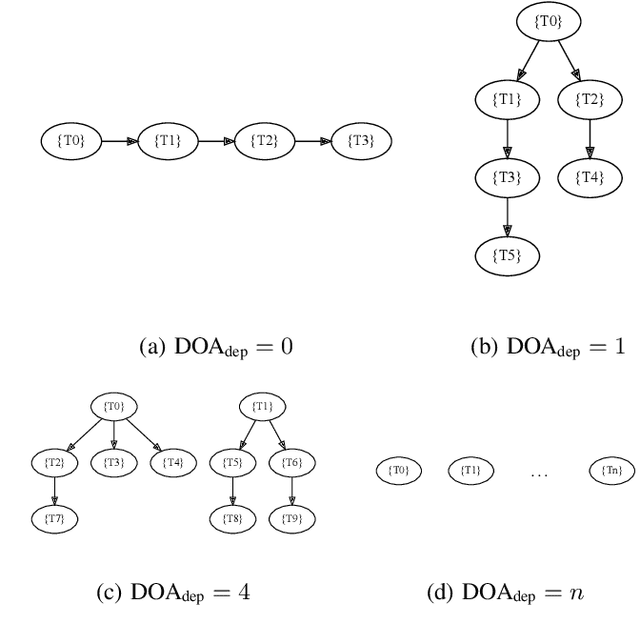
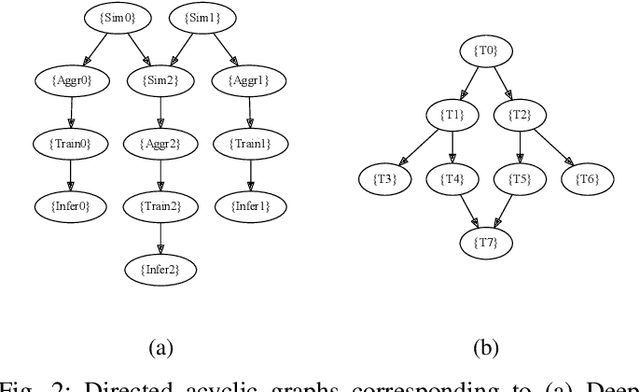
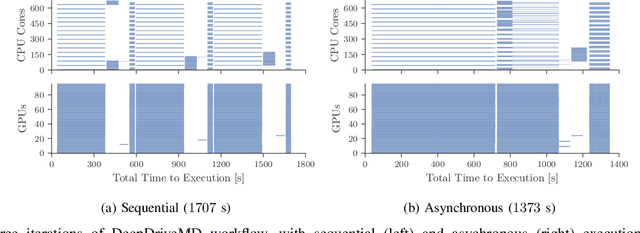
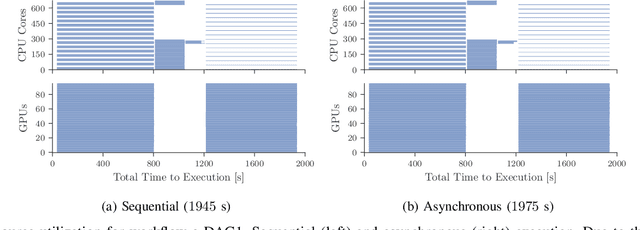
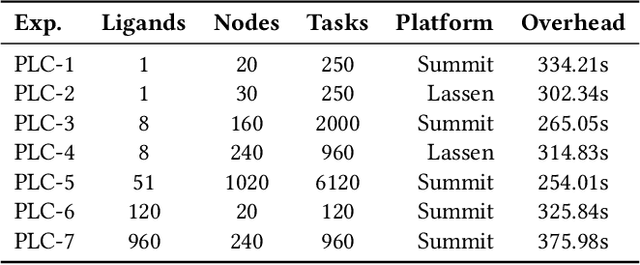
Abstract:Heterogeneous scientific workflows consist of numerous types of tasks and dependencies between them. Middleware capable of scheduling and submitting different task types across heterogeneous platforms must permit asynchronous execution of tasks for improved resource utilization, task throughput, and reduced makespan. In this paper we present an analysis of an important class of heterogeneous workflows, viz., AI-driven HPC workflows, to investigate asynchronous task execution requirements and properties. We model the degree of asynchronicity permitted for arbitrary workflows, and propose key metrics that can be used to determine qualitative benefits when employing asynchronous execution. Our experiments represent important scientific drivers, are performed at scale on Summit, and performance enhancements due to asynchronous execution are consistent with our model.
Protein-Ligand Docking Surrogate Models: A SARS-CoV-2 Benchmark for Deep Learning Accelerated Virtual Screening
Jun 30, 2021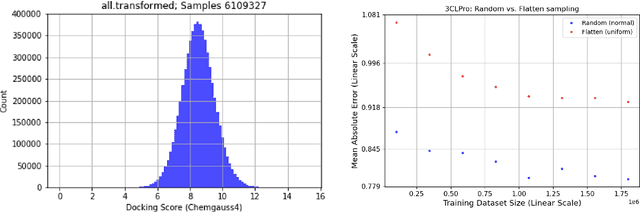
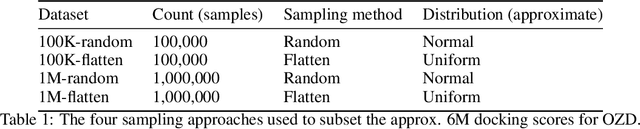
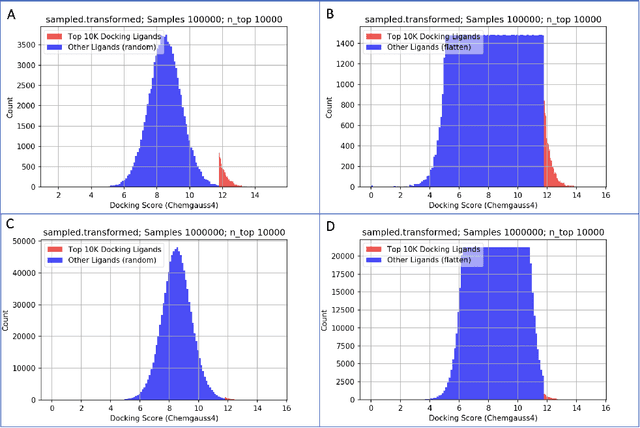
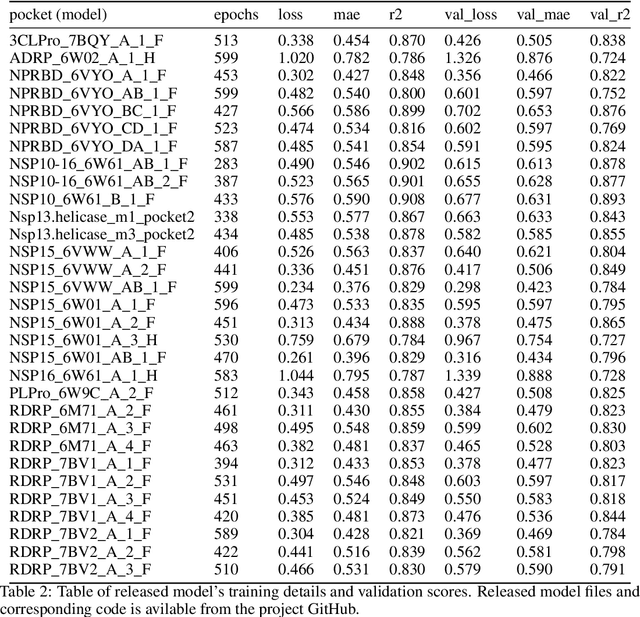
Abstract:We propose a benchmark to study surrogate model accuracy for protein-ligand docking. We share a dataset consisting of 200 million 3D complex structures and 2D structure scores across a consistent set of 13 million "in-stock" molecules over 15 receptors, or binding sites, across the SARS-CoV-2 proteome. Our work shows surrogate docking models have six orders of magnitude more throughput than standard docking protocols on the same supercomputer node types. We demonstrate the power of high-speed surrogate models by running each target against 1 billion molecules in under a day (50k predictions per GPU seconds). We showcase a workflow for docking utilizing surrogate ML models as a pre-filter. Our workflow is ten times faster at screening a library of compounds than the standard technique, with an error rate less than 0.01\% of detecting the underlying best scoring 0.1\% of compounds. Our analysis of the speedup explains that to screen more molecules under a docking paradigm, another order of magnitude speedup must come from model accuracy rather than computing speed (which, if increased, will not anymore alter our throughput to screen molecules). We believe this is strong evidence for the community to begin focusing on improving the accuracy of surrogate models to improve the ability to screen massive compound libraries 100x or even 1000x faster than current techniques.
Achieving 100X faster simulations of complex biological phenomena by coupling ML to HPC ensembles
Apr 26, 2021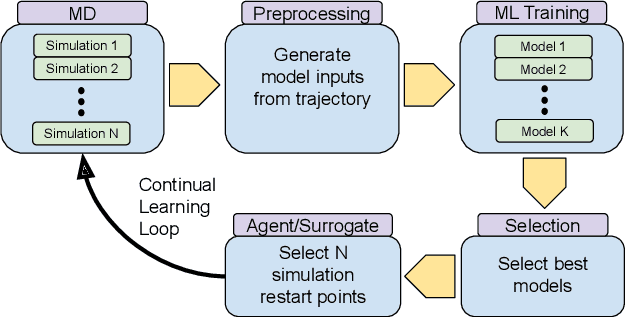
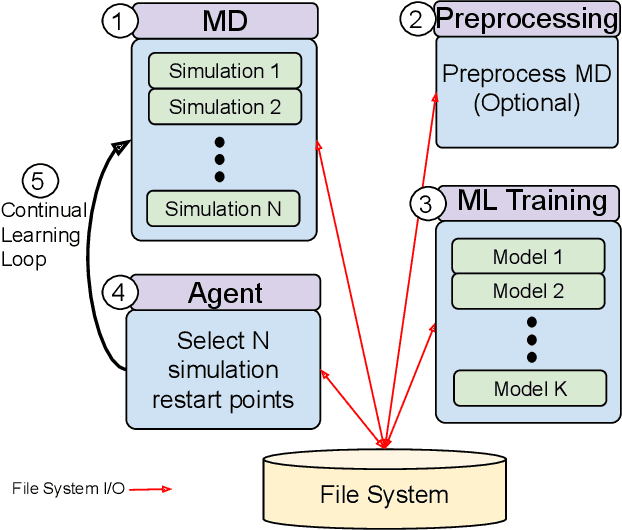
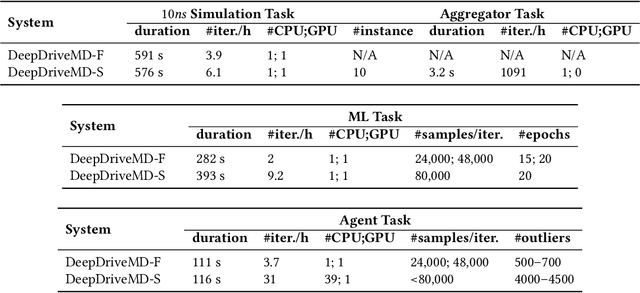
Abstract:The use of ML methods to dynamically steer ensemble-based simulations promises significant improvements in the performance of scientific applications. We present DeepDriveMD, a tool for a range of prototypical ML-driven HPC simulation scenarios, and use it to quantify improvements in the scientific performance of ML-driven ensemble-based applications. We discuss its design and characterize its performance. Motivated by the potential for further scientific improvements and applicability to more sophisticated physical systems, we extend the design of DeepDriveMD to support stream-based communication between simulations and learning methods. It demonstrates a 100x speedup to fold proteins, and performs 1.6x more simulations per unit time, improving resource utilization compared to the sequential framework. Experiments are performed on leadership-class platforms, at scales of up to O(1000) nodes, and for production workloads. We establish DeepDriveMD as a high-performance framework for ML-driven HPC simulation scenarios, that supports diverse simulation and ML back-ends, and which enables new scientific insights by improving length- and time-scale accessed.
Pandemic Drugs at Pandemic Speed: Accelerating COVID-19 Drug Discovery with Hybrid Machine Learning- and Physics-based Simulations on High Performance Computers
Mar 04, 2021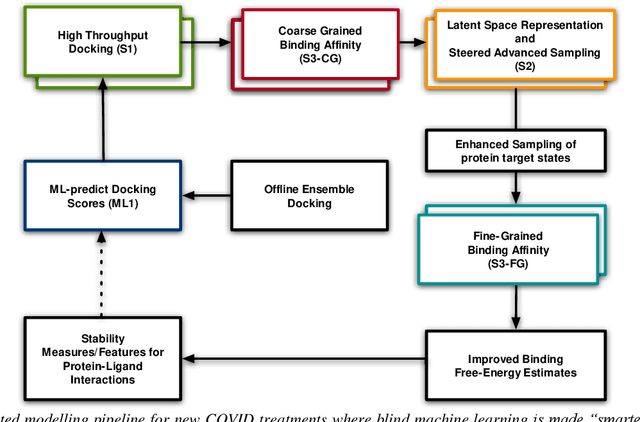
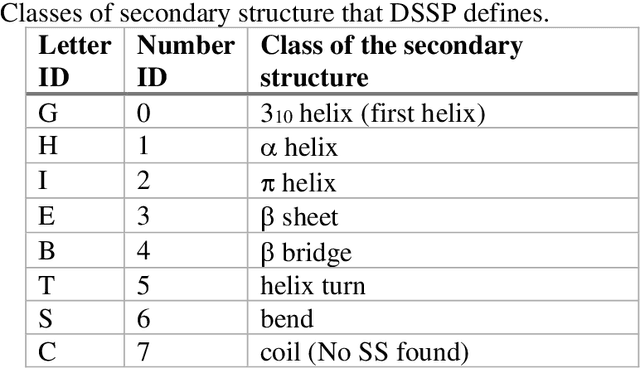
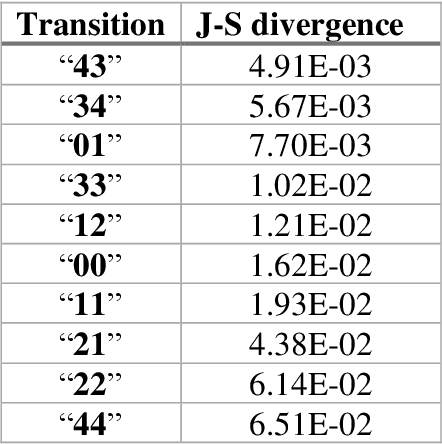
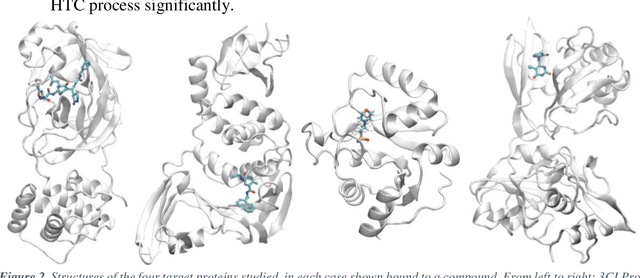
Abstract:The race to meet the challenges of the global pandemic has served as a reminder that the existing drug discovery process is expensive, inefficient and slow. There is a major bottleneck screening the vast number of potential small molecules to shortlist lead compounds for antiviral drug development. New opportunities to accelerate drug discovery lie at the interface between machine learning methods, in this case developed for linear accelerators, and physics-based methods. The two in silico methods, each have their own advantages and limitations which, interestingly, complement each other. Here, we present an innovative method that combines both approaches to accelerate drug discovery. The scale of the resulting workflow is such that it is dependent on high performance computing. We have demonstrated the applicability of this workflow on four COVID-19 target proteins and our ability to perform the required large-scale calculations to identify lead compounds on a variety of supercomputers.
DeepDriveMD: Deep-Learning Driven Adaptive Molecular Simulations for Protein Folding
Sep 17, 2019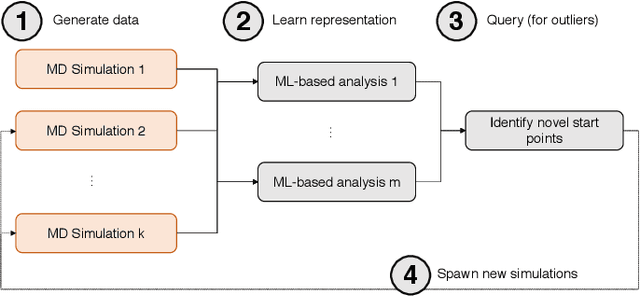
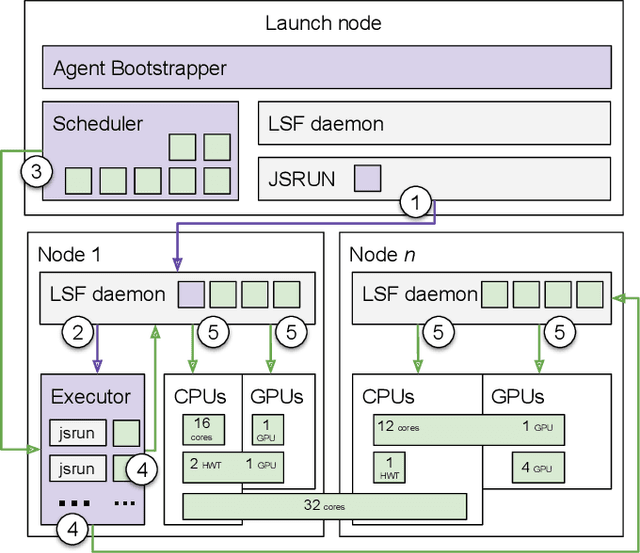
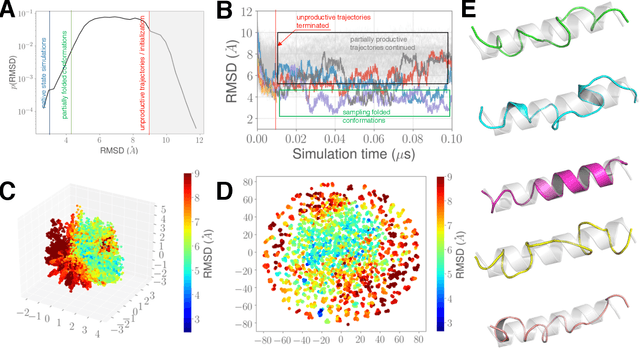
Abstract:Simulations of biological macromolecules play an important role in understanding the physical basis of a number of complex processes such as protein folding. Even with increasing computational power and evolution of specialized architectures, the ability to simulate protein folding at atomistic scales still remains challenging. This stems from the dual aspects of high dimensionality of protein conformational landscapes, and the inability of atomistic molecular dynamics (MD) simulations to sufficiently sample these landscapes to observe folding events. Machine learning/deep learning (ML/DL) techniques, when combined with atomistic MD simulations offer the opportunity to potentially overcome these limitations by: (1) effectively reducing the dimensionality of MD simulations to automatically build latent representations that correspond to biophysically relevant reaction coordinates (RCs), and (2) driving MD simulations to automatically sample potentially novel conformational states based on these RCs. We examine how coupling DL approaches with MD simulations can fold small proteins effectively on supercomputers. In particular, we study the computational costs and effectiveness of scaling DL-coupled MD workflows by folding two prototypical systems, viz., Fs-peptide and the fast-folding variant of the villin head piece protein. We demonstrate that a DL driven MD workflow is able to effectively learn latent representations and drive adaptive simulations. Compared to traditional MD-based approaches, our approach achieves an effective performance gain in sampling the folded states by at least 2.3x. Our study provides a quantitative basis to understand how DL driven MD simulations, can lead to effective performance gains and reduced times to solution on supercomputing resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge