Marta Vallejo
Validating Vision Transformers for Otoscopy: Performance and Data-Leakage Effects
Nov 06, 2025



Abstract:This study evaluates the efficacy of vision transformer models, specifically Swin transformers, in enhancing the diagnostic accuracy of ear diseases compared to traditional convolutional neural networks. With a reported 27% misdiagnosis rate among specialist otolaryngologists, improving diagnostic accuracy is crucial. The research utilised a real-world dataset from the Department of Otolaryngology at the Clinical Hospital of the Universidad de Chile, comprising otoscopic videos of ear examinations depicting various middle and external ear conditions. Frames were selected based on the Laplacian and Shannon entropy thresholds, with blank frames removed. Initially, Swin v1 and Swin v2 transformer models achieved accuracies of 100% and 99.1%, respectively, marginally outperforming the ResNet model (99.5%). These results surpassed metrics reported in related studies. However, the evaluation uncovered a critical data leakage issue in the preprocessing step, affecting both this study and related research using the same raw dataset. After mitigating the data leakage, model performance decreased significantly. Corrected accuracies were 83% for both Swin v1 and Swin v2, and 82% for the ResNet model. This finding highlights the importance of rigorous data handling in machine learning studies, especially in medical applications. The findings indicate that while vision transformers show promise, it is essential to find an optimal balance between the benefits of advanced model architectures and those derived from effective data preprocessing. This balance is key to developing a reliable machine learning model for diagnosing ear diseases.
Deep Learning-Assisted Co-registration of Full-Spectral Autofluorescence Lifetime Microscopic Images with H&E-Stained Histology Images
Feb 15, 2022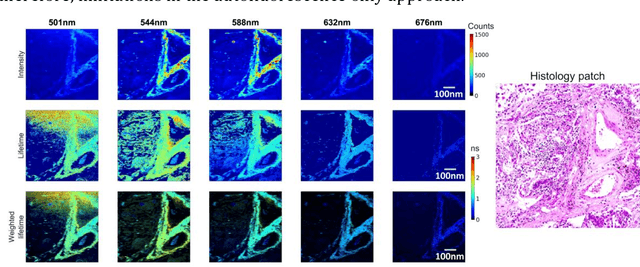
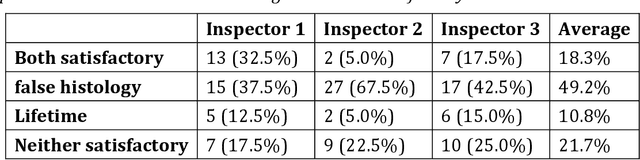
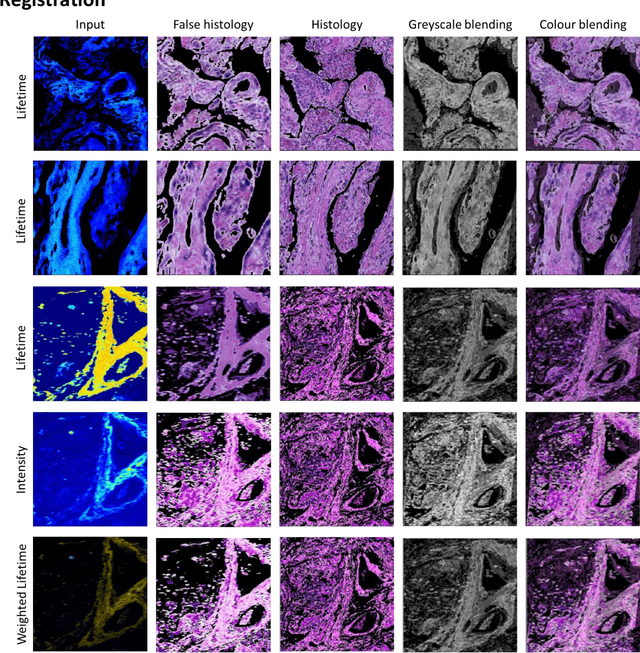
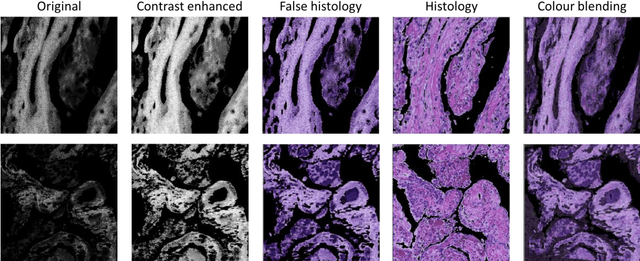
Abstract:Autofluorescence lifetime images reveal unique characteristics of endogenous fluorescence in biological samples. Comprehensive understanding and clinical diagnosis rely on co-registration with the gold standard, histology images, which is extremely challenging due to the difference of both images. Here, we show an unsupervised image-to-image translation network that significantly improves the success of the co-registration using a conventional optimisation-based regression network, applicable to autofluorescence lifetime images at different emission wavelengths. A preliminary blind comparison by experienced researchers shows the superiority of our method on co-registration. The results also indicate that the approach is applicable to various image formats, like fluorescence intensity images. With the registration, stitching outcomes illustrate the distinct differences of the spectral lifetime across an unstained tissue, enabling macro-level rapid visual identification of lung cancer and cellular-level characterisation of cell variants and common types. The approach could be effortlessly extended to lifetime images beyond this range and other staining technologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge