Kian Kenyon-Dean
A Cross Modal Knowledge Distillation & Data Augmentation Recipe for Improving Transcriptomics Representations through Morphological Features
May 27, 2025Abstract:Understanding cellular responses to stimuli is crucial for biological discovery and drug development. Transcriptomics provides interpretable, gene-level insights, while microscopy imaging offers rich predictive features but is harder to interpret. Weakly paired datasets, where samples share biological states, enable multimodal learning but are scarce, limiting their utility for training and multimodal inference. We propose a framework to enhance transcriptomics by distilling knowledge from microscopy images. Using weakly paired data, our method aligns and binds modalities, enriching gene expression representations with morphological information. To address data scarcity, we introduce (1) Semi-Clipped, an adaptation of CLIP for cross-modal distillation using pretrained foundation models, achieving state-of-the-art results, and (2) PEA (Perturbation Embedding Augmentation), a novel augmentation technique that enhances transcriptomics data while preserving inherent biological information. These strategies improve the predictive power and retain the interpretability of transcriptomics, enabling rich unimodal representations for complex biological tasks.
RxRx3-core: Benchmarking drug-target interactions in High-Content Microscopy
Mar 26, 2025



Abstract:High Content Screening (HCS) microscopy datasets have transformed the ability to profile cellular responses to genetic and chemical perturbations, enabling cell-based inference of drug-target interactions (DTI). However, the adoption of representation learning methods for HCS data has been hindered by the lack of accessible datasets and robust benchmarks. To address this gap, we present RxRx3-core, a curated and compressed subset of the RxRx3 dataset, and an associated DTI benchmarking task. At just 18GB, RxRx3-core significantly reduces the size barrier associated with large-scale HCS datasets while preserving critical data necessary for benchmarking representation learning models against a zero-shot DTI prediction task. RxRx3-core includes 222,601 microscopy images spanning 736 CRISPR knockouts and 1,674 compounds at 8 concentrations. RxRx3-core is available on HuggingFace and Polaris, along with pre-trained embeddings and benchmarking code, ensuring accessibility for the research community. By providing a compact dataset and robust benchmarks, we aim to accelerate innovation in representation learning methods for HCS data and support the discovery of novel biological insights.
Towards scientific discovery with dictionary learning: Extracting biological concepts from microscopy foundation models
Dec 20, 2024Abstract:Dictionary learning (DL) has emerged as a powerful interpretability tool for large language models. By extracting known concepts (e.g., Golden-Gate Bridge) from human-interpretable data (e.g., text), sparse DL can elucidate a model's inner workings. In this work, we ask if DL can also be used to discover unknown concepts from less human-interpretable scientific data (e.g., cell images), ultimately enabling modern approaches to scientific discovery. As a first step, we use DL algorithms to study microscopy foundation models trained on multi-cell image data, where little prior knowledge exists regarding which high-level concepts should arise. We show that sparse dictionaries indeed extract biologically-meaningful concepts such as cell type and genetic perturbation type. We also propose a new DL algorithm, Iterative Codebook Feature Learning~(ICFL), and combine it with a pre-processing step that uses PCA whitening from a control dataset. In our experiments, we demonstrate that both ICFL and PCA improve the selectivity of extracted features compared to TopK sparse autoencoders.
ViTally Consistent: Scaling Biological Representation Learning for Cell Microscopy
Nov 04, 2024



Abstract:Large-scale cell microscopy screens are used in drug discovery and molecular biology research to study the effects of millions of chemical and genetic perturbations on cells. To use these images in downstream analysis, we need models that can map each image into a feature space that represents diverse biological phenotypes consistently, in the sense that perturbations with similar biological effects have similar representations. In this work, we present the largest foundation model for cell microscopy data to date, a new 1.9 billion-parameter ViT-G/8 MAE trained on over 8 billion microscopy image crops. Compared to a previous published ViT-L/8 MAE, our new model achieves a 60% improvement in linear separability of genetic perturbations and obtains the best overall performance on whole-genome biological relationship recall and replicate consistency benchmarks. Beyond scaling, we developed two key methods that improve performance: (1) training on a curated and diverse dataset; and, (2) using biologically motivated linear probing tasks to search across each transformer block for the best candidate representation of whole-genome screens. We find that many self-supervised vision transformers, pretrained on either natural or microscopy images, yield significantly more biologically meaningful representations of microscopy images in their intermediate blocks than in their typically used final blocks. More broadly, our approach and results provide insights toward a general strategy for successfully building foundation models for large-scale biological data.
Benchmarking Transcriptomics Foundation Models for Perturbation Analysis : one PCA still rules them all
Oct 17, 2024



Abstract:Understanding the relationships among genes, compounds, and their interactions in living organisms remains limited due to technological constraints and the complexity of biological data. Deep learning has shown promise in exploring these relationships using various data types. However, transcriptomics, which provides detailed insights into cellular states, is still underused due to its high noise levels and limited data availability. Recent advancements in transcriptomics sequencing provide new opportunities to uncover valuable insights, especially with the rise of many new foundation models for transcriptomics, yet no benchmark has been made to robustly evaluate the effectiveness of these rising models for perturbation analysis. This article presents a novel biologically motivated evaluation framework and a hierarchy of perturbation analysis tasks for comparing the performance of pretrained foundation models to each other and to more classical techniques of learning from transcriptomics data. We compile diverse public datasets from different sequencing techniques and cell lines to assess models performance. Our approach identifies scVI and PCA to be far better suited models for understanding biological perturbations in comparison to existing foundation models, especially in their application in real-world scenarios.
Masked Autoencoders for Microscopy are Scalable Learners of Cellular Biology
Apr 16, 2024



Abstract:Featurizing microscopy images for use in biological research remains a significant challenge, especially for large-scale experiments spanning millions of images. This work explores the scaling properties of weakly supervised classifiers and self-supervised masked autoencoders (MAEs) when training with increasingly larger model backbones and microscopy datasets. Our results show that ViT-based MAEs outperform weakly supervised classifiers on a variety of tasks, achieving as much as a 11.5% relative improvement when recalling known biological relationships curated from public databases. Additionally, we develop a new channel-agnostic MAE architecture (CA-MAE) that allows for inputting images of different numbers and orders of channels at inference time. We demonstrate that CA-MAEs effectively generalize by inferring and evaluating on a microscopy image dataset (JUMP-CP) generated under different experimental conditions with a different channel structure than our pretraining data (RPI-93M). Our findings motivate continued research into scaling self-supervised learning on microscopy data in order to create powerful foundation models of cellular biology that have the potential to catalyze advancements in drug discovery and beyond.
Masked autoencoders are scalable learners of cellular morphology
Sep 27, 2023



Abstract:Inferring biological relationships from cellular phenotypes in high-content microscopy screens provides significant opportunity and challenge in biological research. Prior results have shown that deep vision models can capture biological signal better than hand-crafted features. This work explores how weakly supervised and self-supervised deep learning approaches scale when training larger models on larger datasets. Our results show that both CNN- and ViT-based masked autoencoders significantly outperform weakly supervised models. At the high-end of our scale, a ViT-L/8 trained on over 3.5-billion unique crops sampled from 95-million microscopy images achieves relative improvements as high as 28% over our best weakly supervised models at inferring known biological relationships curated from public databases.
Deconstructing word embedding algorithms
Nov 12, 2020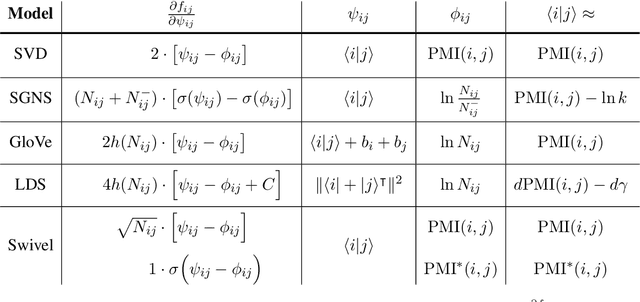
Abstract:Word embeddings are reliable feature representations of words used to obtain high quality results for various NLP applications. Uncontextualized word embeddings are used in many NLP tasks today, especially in resource-limited settings where high memory capacity and GPUs are not available. Given the historical success of word embeddings in NLP, we propose a retrospective on some of the most well-known word embedding algorithms. In this work, we deconstruct Word2vec, GloVe, and others, into a common form, unveiling some of the common conditions that seem to be required for making performant word embeddings. We believe that the theoretical findings in this paper can provide a basis for more informed development of future models.
Learning Efficient Task-Specific Meta-Embeddings with Word Prisms
Nov 05, 2020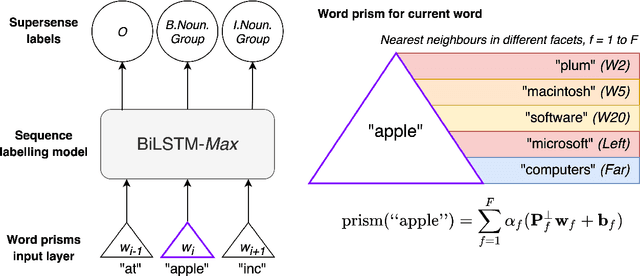
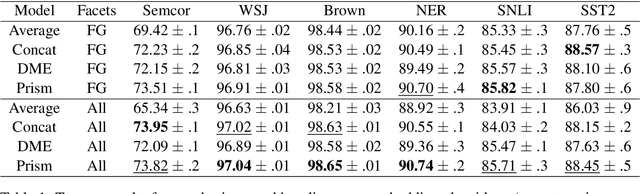
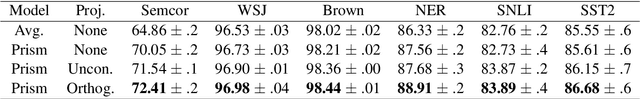
Abstract:Word embeddings are trained to predict word cooccurrence statistics, which leads them to possess different lexical properties (syntactic, semantic, etc.) depending on the notion of context defined at training time. These properties manifest when querying the embedding space for the most similar vectors, and when used at the input layer of deep neural networks trained to solve downstream NLP problems. Meta-embeddings combine multiple sets of differently trained word embeddings, and have been shown to successfully improve intrinsic and extrinsic performance over equivalent models which use just one set of source embeddings. We introduce word prisms: a simple and efficient meta-embedding method that learns to combine source embeddings according to the task at hand. Word prisms learn orthogonal transformations to linearly combine the input source embeddings, which allows them to be very efficient at inference time. We evaluate word prisms in comparison to other meta-embedding methods on six extrinsic evaluations and observe that word prisms offer improvements in performance on all tasks.
Deconstructing and reconstructing word embedding algorithms
Nov 29, 2019
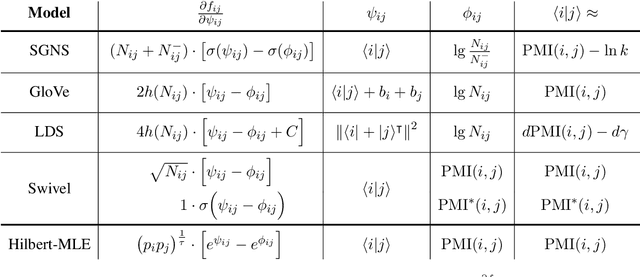
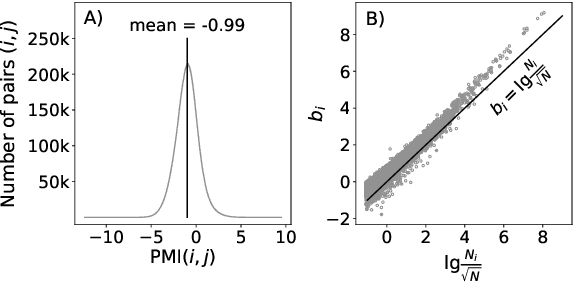
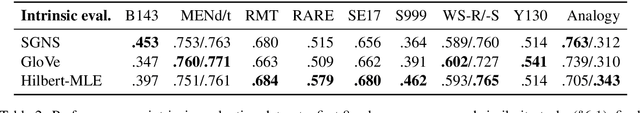
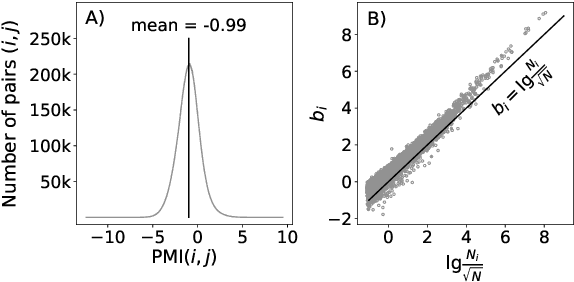
Abstract:Uncontextualized word embeddings are reliable feature representations of words used to obtain high quality results for various NLP applications. Given the historical success of word embeddings in NLP, we propose a retrospective on some of the most well-known word embedding algorithms. In this work, we deconstruct Word2vec, GloVe, and others, into a common form, unveiling some of the necessary and sufficient conditions required for making performant word embeddings. We find that each algorithm: (1) fits vector-covector dot products to approximate pointwise mutual information (PMI); and, (2) modulates the loss gradient to balance weak and strong signals. We demonstrate that these two algorithmic features are sufficient conditions to construct a novel word embedding algorithm, Hilbert-MLE. We find that its embeddings obtain equivalent or better performance against other algorithms across 17 intrinsic and extrinsic datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge