Kevin Kröninger
Efficient Complex-Valued Vision Transformers for MRI Classification Directly from k-Space
Jan 26, 2026Abstract:Deep learning applications in Magnetic Resonance Imaging (MRI) predominantly operate on reconstructed magnitude images, a process that discards phase information and requires computationally expensive transforms. Standard neural network architectures rely on local operations (convolutions or grid-patches) that are ill-suited for the global, non-local nature of raw frequency-domain (k-Space) data. In this work, we propose a novel complex-valued Vision Transformer (kViT) designed to perform classification directly on k-Space data. To bridge the geometric disconnect between current architectures and MRI physics, we introduce a radial k-Space patching strategy that respects the spectral energy distribution of the frequency-domain. Extensive experiments on the fastMRI and in-house datasets demonstrate that our approach achieves classification performance competitive with state-of-the-art image-domain baselines (ResNet, EfficientNet, ViT). Crucially, kViT exhibits superior robustness to high acceleration factors and offers a paradigm shift in computational efficiency, reducing VRAM consumption during training by up to 68$\times$ compared to standard methods. This establishes a pathway for resource-efficient, direct-from-scanner AI analysis.
PhaseGen: A Diffusion-Based Approach for Complex-Valued MRI Data Generation
Apr 10, 2025Abstract:Magnetic resonance imaging (MRI) raw data, or k-Space data, is complex-valued, containing both magnitude and phase information. However, clinical and existing Artificial Intelligence (AI)-based methods focus only on magnitude images, discarding the phase data despite its potential for downstream tasks, such as tumor segmentation and classification. In this work, we introduce $\textit{PhaseGen}$, a novel complex-valued diffusion model for generating synthetic MRI raw data conditioned on magnitude images, commonly used in clinical practice. This enables the creation of artificial complex-valued raw data, allowing pretraining for models that require k-Space information. We evaluate PhaseGen on two tasks: skull-stripping directly in k-Space and MRI reconstruction using the publicly available FastMRI dataset. Our results show that training with synthetic phase data significantly improves generalization for skull-stripping on real-world data, with an increased segmentation accuracy from $41.1\%$ to $80.1\%$, and enhances MRI reconstruction when combined with limited real-world data. This work presents a step forward in utilizing generative AI to bridge the gap between magnitude-based datasets and the complex-valued nature of MRI raw data. This approach allows researchers to leverage the vast amount of avaliable image domain data in combination with the information-rich k-Space data for more accurate and efficient diagnostic tasks. We make our code publicly $\href{https://github.com/TIO-IKIM/PhaseGen}{\text{available here}}$.
k-strip: A novel segmentation algorithm in k-space for the application of skull stripping
May 19, 2022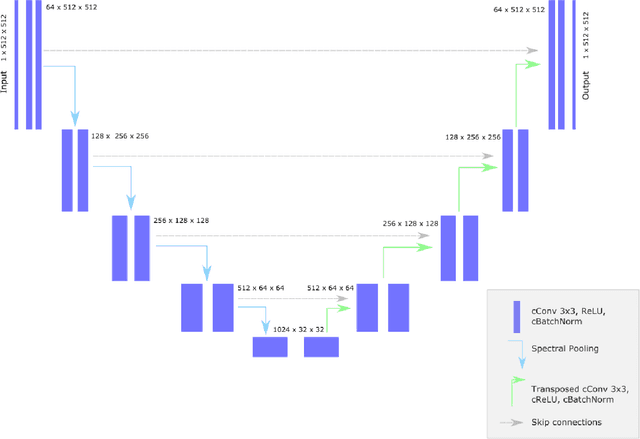
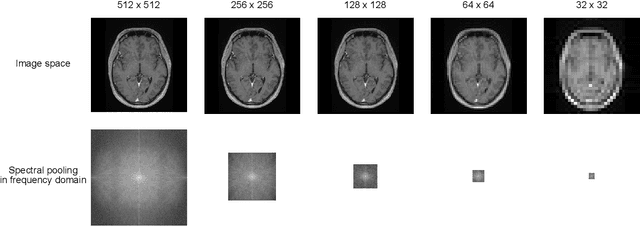
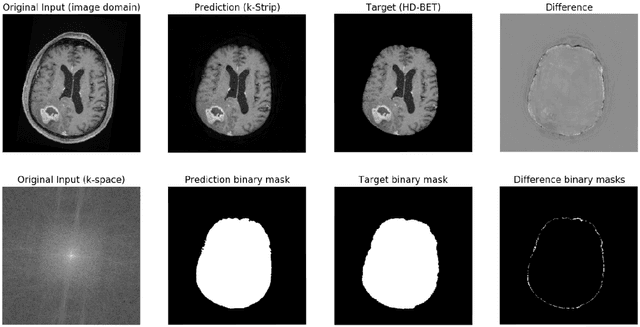
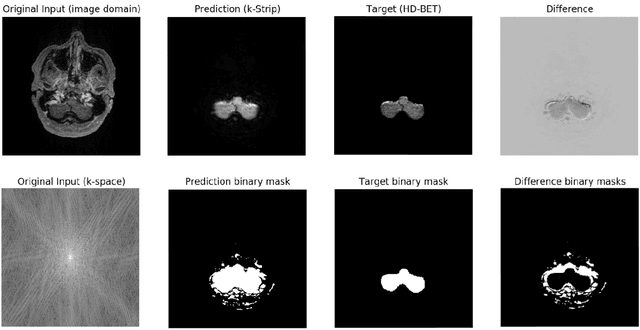
Abstract:Objectives: Present a novel deep learning-based skull stripping algorithm for magnetic resonance imaging (MRI) that works directly in the information rich k-space. Materials and Methods: Using two datasets from different institutions with a total of 36,900 MRI slices, we trained a deep learning-based model to work directly with the complex raw k-space data. Skull stripping performed by HD-BET (Brain Extraction Tool) in the image domain were used as the ground truth. Results: Both datasets were very similar to the ground truth (DICE scores of 92\%-98\% and Hausdorff distances of under 5.5 mm). Results on slices above the eye-region reach DICE scores of up to 99\%, while the accuracy drops in regions around the eyes and below, with partially blurred output. The output of k-strip often smoothed edges at the demarcation to the skull. Binary masks are created with an appropriate threshold. Conclusion: With this proof-of-concept study, we were able to show the feasibility of working in the k-space frequency domain, preserving phase information, with consistent results. Future research should be dedicated to discovering additional ways the k-space can be used for innovative image analysis and further workflows.
BAT.jl -- A Julia-based tool for Bayesian inference
Aug 07, 2020



Abstract:We describe the development of a multi-purpose software for Bayesian statistical inference, BAT.jl, written in the Julia language. The major design considerations and implemented algorithms are summarized here, together with a test suite that ensures the proper functioning of the algorithms. We also give an extended example from the realm of physics that demonstrates the functionalities of BAT.jl.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge