Jonathan R. Dillman
Can Modern NLP Systems Reliably Annotate Chest Radiography Exams? A Pre-Purchase Evaluation and Comparative Study of Solutions from AWS, Google, Azure, John Snow Labs, and Open-Source Models on an Independent Pediatric Dataset
May 29, 2025Abstract:General-purpose clinical natural language processing (NLP) tools are increasingly used for the automatic labeling of clinical reports. However, independent evaluations for specific tasks, such as pediatric chest radiograph (CXR) report labeling, are limited. This study compares four commercial clinical NLP systems - Amazon Comprehend Medical (AWS), Google Healthcare NLP (GC), Azure Clinical NLP (AZ), and SparkNLP (SP) - for entity extraction and assertion detection in pediatric CXR reports. Additionally, CheXpert and CheXbert, two dedicated chest radiograph report labelers, were evaluated on the same task using CheXpert-defined labels. We analyzed 95,008 pediatric CXR reports from a large academic pediatric hospital. Entities and assertion statuses (positive, negative, uncertain) from the findings and impression sections were extracted by the NLP systems, with impression section entities mapped to 12 disease categories and a No Findings category. CheXpert and CheXbert extracted the same 13 categories. Outputs were compared using Fleiss Kappa and accuracy against a consensus pseudo-ground truth. Significant differences were found in the number of extracted entities and assertion distributions across NLP systems. SP extracted 49,688 unique entities, GC 16,477, AZ 31,543, and AWS 27,216. Assertion accuracy across models averaged around 62%, with SP highest (76%) and AWS lowest (50%). CheXpert and CheXbert achieved 56% accuracy. Considerable variability in performance highlights the need for careful validation and review before deploying NLP tools for clinical report labeling.
Joint Self-Supervised and Supervised Contrastive Learning for Multimodal MRI Data: Towards Predicting Abnormal Neurodevelopment
Dec 22, 2023



Abstract:The integration of different imaging modalities, such as structural, diffusion tensor, and functional magnetic resonance imaging, with deep learning models has yielded promising outcomes in discerning phenotypic characteristics and enhancing disease diagnosis. The development of such a technique hinges on the efficient fusion of heterogeneous multimodal features, which initially reside within distinct representation spaces. Naively fusing the multimodal features does not adequately capture the complementary information and could even produce redundancy. In this work, we present a novel joint self-supervised and supervised contrastive learning method to learn the robust latent feature representation from multimodal MRI data, allowing the projection of heterogeneous features into a shared common space, and thereby amalgamating both complementary and analogous information across various modalities and among similar subjects. We performed a comparative analysis between our proposed method and alternative deep multimodal learning approaches. Through extensive experiments on two independent datasets, the results demonstrated that our method is significantly superior to several other deep multimodal learning methods in predicting abnormal neurodevelopment. Our method has the capability to facilitate computer-aided diagnosis within clinical practice, harnessing the power of multimodal data.
A Novel Collaborative Self-Supervised Learning Method for Radiomic Data
Feb 20, 2023



Abstract:The computer-aided disease diagnosis from radiomic data is important in many medical applications. However, developing such a technique relies on annotating radiological images, which is a time-consuming, labor-intensive, and expensive process. In this work, we present the first novel collaborative self-supervised learning method to solve the challenge of insufficient labeled radiomic data, whose characteristics are different from text and image data. To achieve this, we present two collaborative pretext tasks that explore the latent pathological or biological relationships between regions of interest and the similarity and dissimilarity information between subjects. Our method collaboratively learns the robust latent feature representations from radiomic data in a self-supervised manner to reduce human annotation efforts, which benefits the disease diagnosis. We compared our proposed method with other state-of-the-art self-supervised learning methods on a simulation study and two independent datasets. Extensive experimental results demonstrated that our method outperforms other self-supervised learning methods on both classification and regression tasks. With further refinement, our method shows the potential advantage in automatic disease diagnosis with large-scale unlabeled data available.
A Novel Ontology-guided Attribute Partitioning Ensemble Learning Model for Early Prediction of Cognitive Deficits using Quantitative Structural MRI in Very Preterm Infants
Feb 08, 2022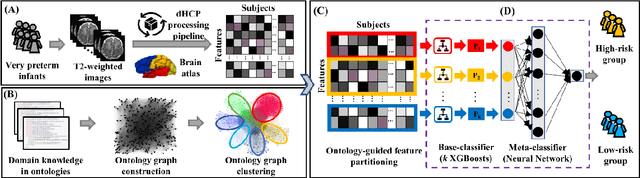
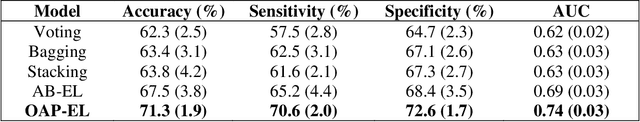
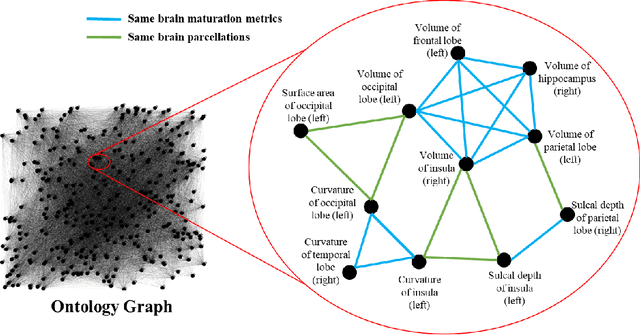
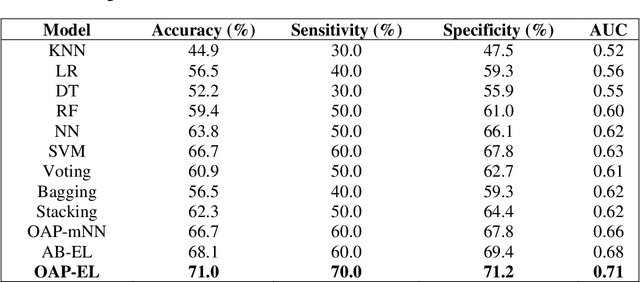
Abstract:Structural magnetic resonance imaging studies have shown that brain anatomical abnormalities are associated with cognitive deficits in preterm infants. Brain maturation and geometric features can be used with machine learning models for predicting later neurodevelopmental deficits. However, traditional machine learning models would suffer from a large feature-to-instance ratio (i.e., a large number of features but a small number of instances/samples). Ensemble learning is a paradigm that strategically generates and integrates a library of machine learning classifiers and has been successfully used on a wide variety of predictive modeling problems to boost model performance. Attribute (i.e., feature) bagging method is the most commonly used feature partitioning scheme, which randomly and repeatedly draws feature subsets from the entire feature set. Although attribute bagging method can effectively reduce feature dimensionality to handle the large feature-to-instance ratio, it lacks consideration of domain knowledge and latent relationship among features. In this study, we proposed a novel Ontology-guided Attribute Partitioning (OAP) method to better draw feature subsets by considering domain-specific relationship among features. With the better partitioned feature subsets, we developed an ensemble learning framework, which is referred to as OAP Ensemble Learning (OAP-EL). We applied the OAP-EL to predict cognitive deficits at 2 year of age using quantitative brain maturation and geometric features obtained at term equivalent age in very preterm infants. We demonstrated that the proposed OAP-EL approach significantly outperformed the peer ensemble learning and traditional machine learning approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge