Jon Duke
The GenAI Generation: Student Views of Awareness, Preparedness, and Concern
May 04, 2025Abstract:Generative AI (GenAI) is revolutionizing education and workforce development, profoundly shaping how students learn, engage, and prepare for their future. Outpacing the development of uniform policies and structures, GenAI has heralded a unique era and given rise to the GenAI Generation: a cohort of students whose education has been increasingly shaped by the opportunities and challenges GenAI presents during its widespread adoption within society. This study examines our students' perceptions of GenAI through a concise survey with optional open-ended questions, focusing on their awareness, preparedness, and concerns. Evaluation of more than 250 responses with more than 40% providing detailed qualitative feedback reveals a core dual sentiment: while most students express enthusiasm for GenAI, an even greater proportion voice a spectrum of concerns about ethics, job displacement, and the adequacy of educational structures given the highly transformative technology. These findings offer critical insights into how students view the potential and pitfalls of GenAI for future career impacts, with accompanying recommendations to guide educational institutions in navigating a future driven by GenAI.
EVA: Generating Longitudinal Electronic Health Records Using Conditional Variational Autoencoders
Dec 18, 2020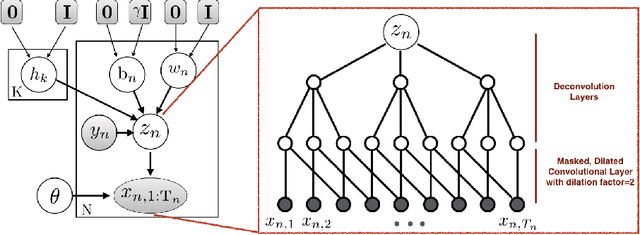
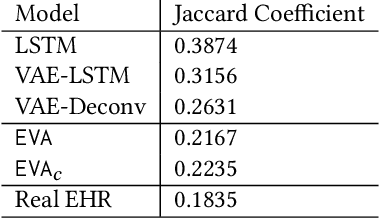
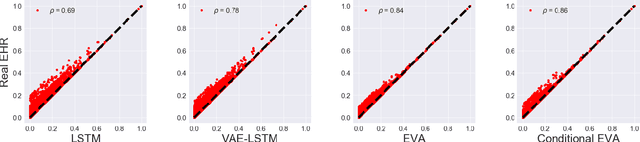
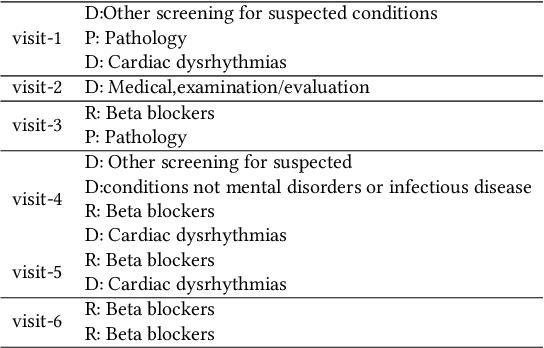
Abstract:Researchers require timely access to real-world longitudinal electronic health records (EHR) to develop, test, validate, and implement machine learning solutions that improve the quality and efficiency of healthcare. In contrast, health systems value deeply patient privacy and data security. De-identified EHRs do not adequately address the needs of health systems, as de-identified data are susceptible to re-identification and its volume is also limited. Synthetic EHRs offer a potential solution. In this paper, we propose EHR Variational Autoencoder (EVA) for synthesizing sequences of discrete EHR encounters (e.g., clinical visits) and encounter features (e.g., diagnoses, medications, procedures). We illustrate that EVA can produce realistic EHR sequences, account for individual differences among patients, and can be conditioned on specific disease conditions, thus enabling disease-specific studies. We design efficient, accurate inference algorithms by combining stochastic gradient Markov Chain Monte Carlo with amortized variational inference. We assess the utility of the methods on large real-world EHR repositories containing over 250, 000 patients. Our experiments, which include user studies with knowledgeable clinicians, indicate the generated EHR sequences are realistic. We confirmed the performance of predictive models trained on the synthetic data are similar with those trained on real EHRs. Additionally, our findings indicate that augmenting real data with synthetic EHRs results in the best predictive performance - improving the best baseline by as much as 8% in top-20 recall.
Explainable Prediction of Medical Codes from Clinical Text
Apr 16, 2018
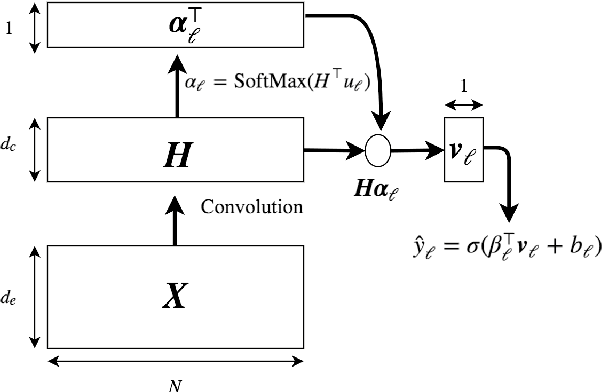
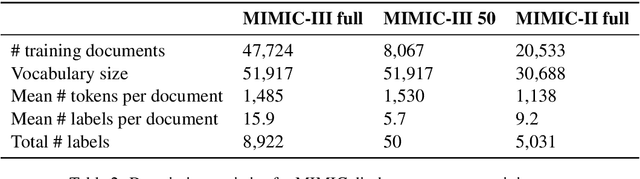
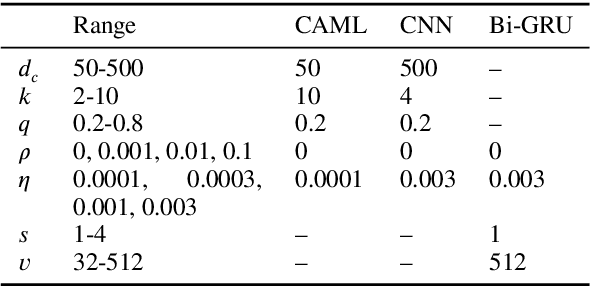
Abstract:Clinical notes are text documents that are created by clinicians for each patient encounter. They are typically accompanied by medical codes, which describe the diagnosis and treatment. Annotating these codes is labor intensive and error prone; furthermore, the connection between the codes and the text is not annotated, obscuring the reasons and details behind specific diagnoses and treatments. We present an attentional convolutional network that predicts medical codes from clinical text. Our method aggregates information across the document using a convolutional neural network, and uses an attention mechanism to select the most relevant segments for each of the thousands of possible codes. The method is accurate, achieving precision@8 of 0.71 and a Micro-F1 of 0.54, which are both better than the prior state of the art. Furthermore, through an interpretability evaluation by a physician, we show that the attention mechanism identifies meaningful explanations for each code assignment
Generating Multi-label Discrete Patient Records using Generative Adversarial Networks
Jan 11, 2018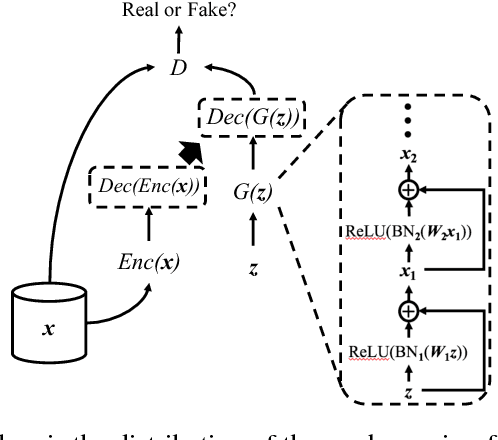
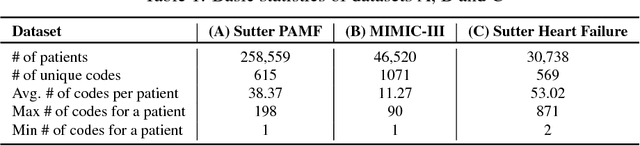
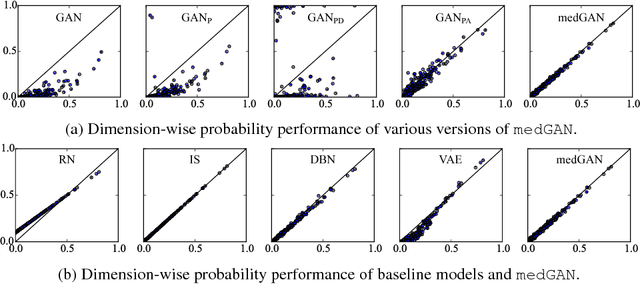
Abstract:Access to electronic health record (EHR) data has motivated computational advances in medical research. However, various concerns, particularly over privacy, can limit access to and collaborative use of EHR data. Sharing synthetic EHR data could mitigate risk. In this paper, we propose a new approach, medical Generative Adversarial Network (medGAN), to generate realistic synthetic patient records. Based on input real patient records, medGAN can generate high-dimensional discrete variables (e.g., binary and count features) via a combination of an autoencoder and generative adversarial networks. We also propose minibatch averaging to efficiently avoid mode collapse, and increase the learning efficiency with batch normalization and shortcut connections. To demonstrate feasibility, we showed that medGAN generates synthetic patient records that achieve comparable performance to real data on many experiments including distribution statistics, predictive modeling tasks and a medical expert review. We also empirically observe a limited privacy risk in both identity and attribute disclosure using medGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge