John W. Garrett
From Embeddings to Accuracy: Comparing Foundation Models for Radiographic Classification
May 16, 2025Abstract:Foundation models, pretrained on extensive datasets, have significantly advanced machine learning by providing robust and transferable embeddings applicable to various domains, including medical imaging diagnostics. This study evaluates the utility of embeddings derived from both general-purpose and medical domain-specific foundation models for training lightweight adapter models in multi-class radiography classification, focusing specifically on tube placement assessment. A dataset comprising 8842 radiographs classified into seven distinct categories was employed to extract embeddings using six foundation models: DenseNet121, BiomedCLIP, Med-Flamingo, MedImageInsight, Rad-DINO, and CXR-Foundation. Adapter models were subsequently trained using classical machine learning algorithms. Among these combinations, MedImageInsight embeddings paired with an support vector machine adapter yielded the highest mean area under the curve (mAUC) at 93.8%, followed closely by Rad-DINO (91.1%) and CXR-Foundation (89.0%). In comparison, BiomedCLIP and DenseNet121 exhibited moderate performance with mAUC scores of 83.0% and 81.8%, respectively, whereas Med-Flamingo delivered the lowest performance at 75.1%. Notably, most adapter models demonstrated computational efficiency, achieving training within one minute and inference within seconds on CPU, underscoring their practicality for clinical applications. Furthermore, fairness analyses on adapters trained on MedImageInsight-derived embeddings indicated minimal disparities, with gender differences in performance within 2% and standard deviations across age groups not exceeding 3%. These findings confirm that foundation model embeddings-especially those from MedImageInsight-facilitate accurate, computationally efficient, and equitable diagnostic classification using lightweight adapters for radiographic image analysis.
A Generalizable Artificial Intelligence Model for COVID-19 Classification Task Using Chest X-ray Radiographs: Evaluated Over Four Clinical Datasets with 15,097 Patients
Oct 04, 2022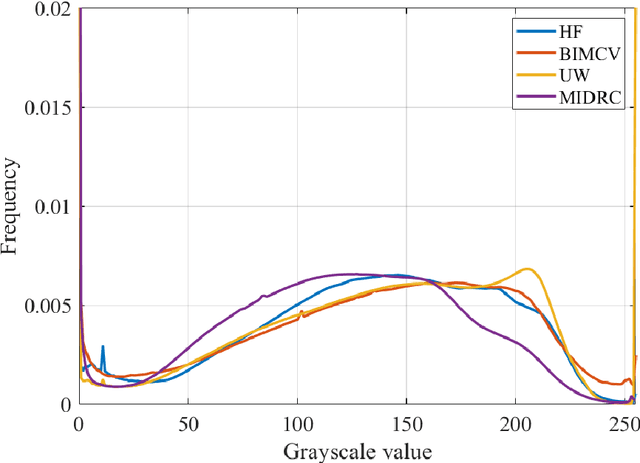
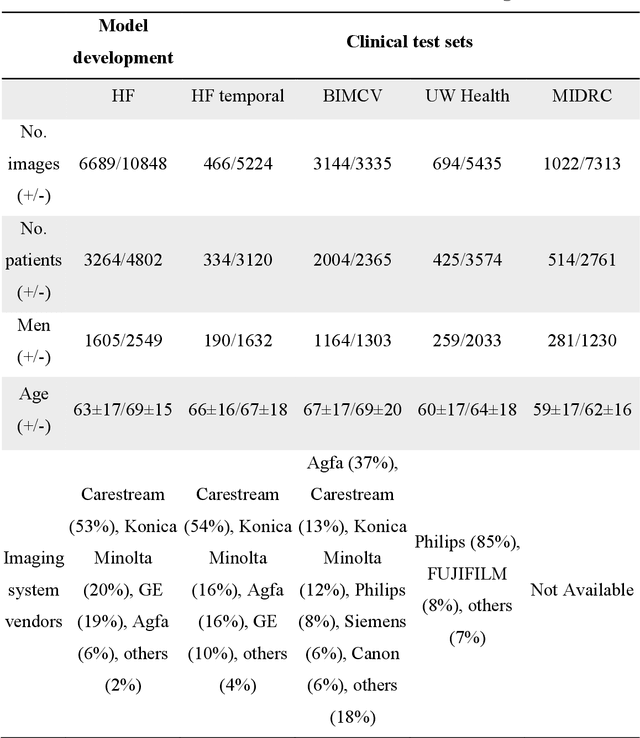
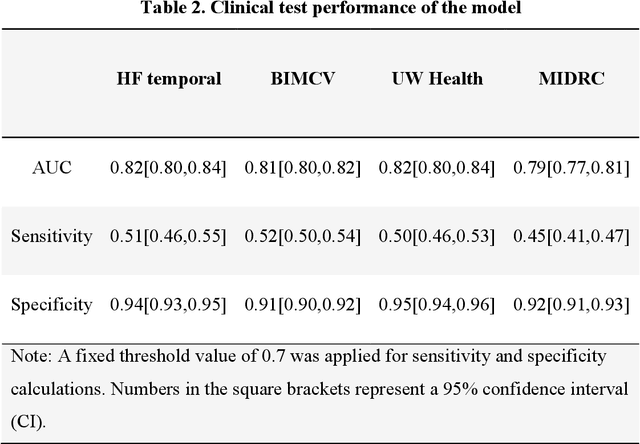
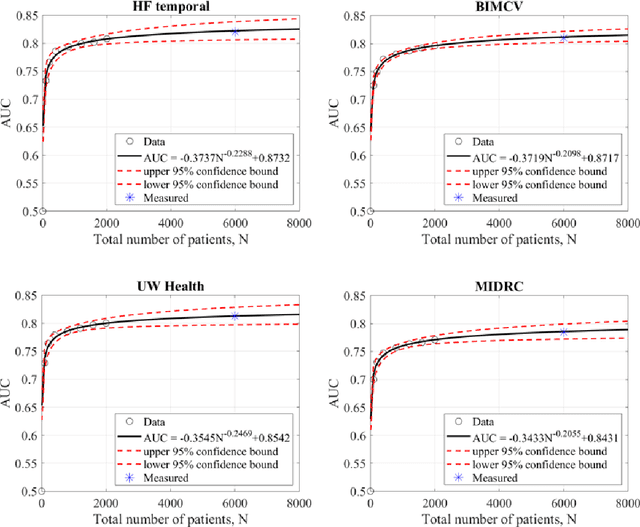
Abstract:Purpose: To answer the long-standing question of whether a model trained from a single clinical site can be generalized to external sites. Materials and Methods: 17,537 chest x-ray radiographs (CXRs) from 3,264 COVID-19-positive patients and 4,802 COVID-19-negative patients were collected from a single site for AI model development. The generalizability of the trained model was retrospectively evaluated using four different real-world clinical datasets with a total of 26,633 CXRs from 15,097 patients (3,277 COVID-19-positive patients). The area under the receiver operating characteristic curve (AUC) was used to assess diagnostic performance. Results: The AI model trained using a single-source clinical dataset achieved an AUC of 0.82 (95% CI: 0.80, 0.84) when applied to the internal temporal test set. When applied to datasets from two external clinical sites, an AUC of 0.81 (95% CI: 0.80, 0.82) and 0.82 (95% CI: 0.80, 0.84) were achieved. An AUC of 0.79 (95% CI: 0.77, 0.81) was achieved when applied to a multi-institutional COVID-19 dataset collected by the Medical Imaging and Data Resource Center (MIDRC). A power-law dependence, N^(k )(k is empirically found to be -0.21 to -0.25), indicates a relatively weak performance dependence on the training data sizes. Conclusion: COVID-19 classification AI model trained using well-curated data from a single clinical site is generalizable to external clinical sites without a significant drop in performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge