Jens Rittscher
Efficient video indexing for monitoring disease activity and progression in the upper gastrointestinal tract
May 10, 2019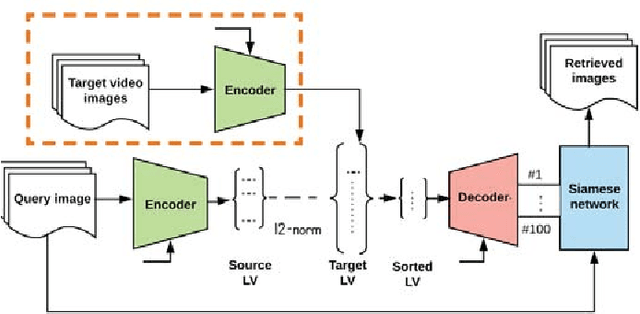
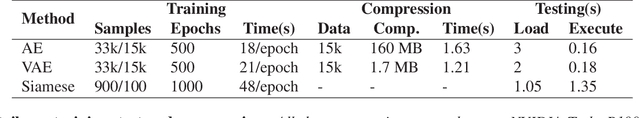
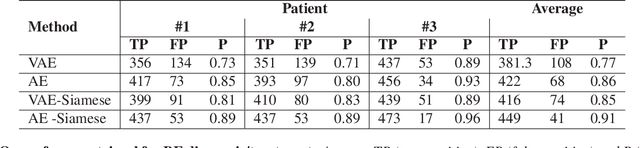
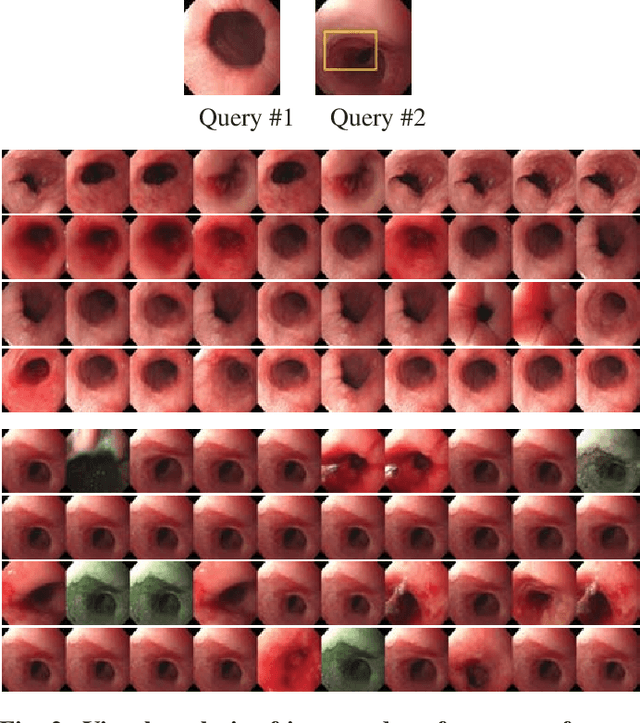
Abstract:Endoscopy is a routine imaging technique used for both diagnosis and minimally invasive surgical treatment. While the endoscopy video contains a wealth of information, tools to capture this information for the purpose of clinical reporting are rather poor. In date, endoscopists do not have any access to tools that enable them to browse the video data in an efficient and user friendly manner. Fast and reliable video retrieval methods could for example, allow them to review data from previous exams and therefore improve their ability to monitor disease progression. Deep learning provides new avenues of compressing and indexing video in an extremely efficient manner. In this study, we propose to use an autoencoder for efficient video compression and fast retrieval of video images. To boost the accuracy of video image retrieval and to address data variability like multi-modality and view-point changes, we propose the integration of a Siamese network. We demonstrate that our approach is competitive in retrieving images from 3 large scale videos of 3 different patients obtained against the query samples of their previous diagnosis. Quantitative validation shows that the combined approach yield an overall improvement of 5% and 8% over classical and variational autoencoders, respectively.
Endoscopy artifact detection (EAD 2019) challenge dataset
May 08, 2019
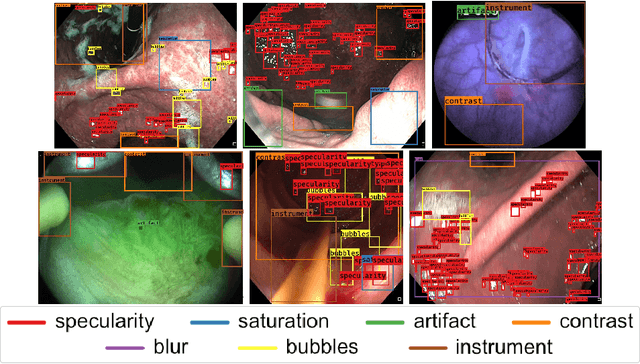
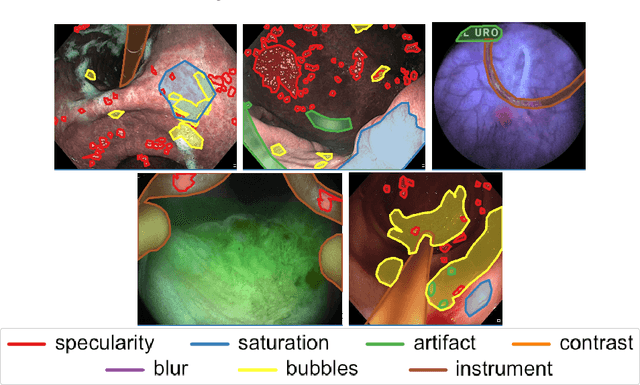
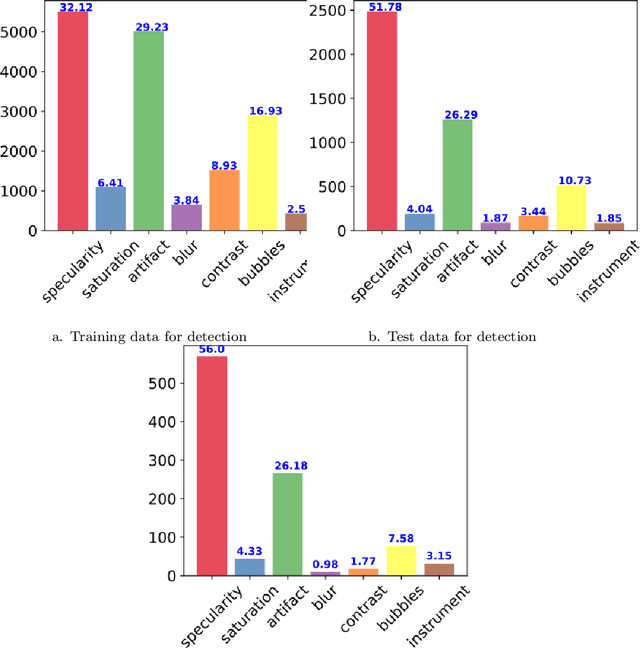
Abstract:Endoscopic artifacts are a core challenge in facilitating the diagnosis and treatment of diseases in hollow organs. Precise detection of specific artifacts like pixel saturations, motion blur, specular reflections, bubbles and debris is essential for high-quality frame restoration and is crucial for realizing reliable computer-assisted tools for improved patient care. At present most videos in endoscopy are currently not analyzed due to the abundant presence of multi-class artifacts in video frames. Through the endoscopic artifact detection (EAD 2019) challenge, we address this key bottleneck problem by solving the accurate identification and localization of endoscopic frame artifacts to enable further key quantitative analysis of unusable video frames such as mosaicking and 3D reconstruction which is crucial for delivering improved patient care. This paper summarizes the challenge tasks and describes the dataset and evaluation criteria established in the EAD 2019 challenge.
A deep learning framework for quality assessment and restoration in video endoscopy
Apr 15, 2019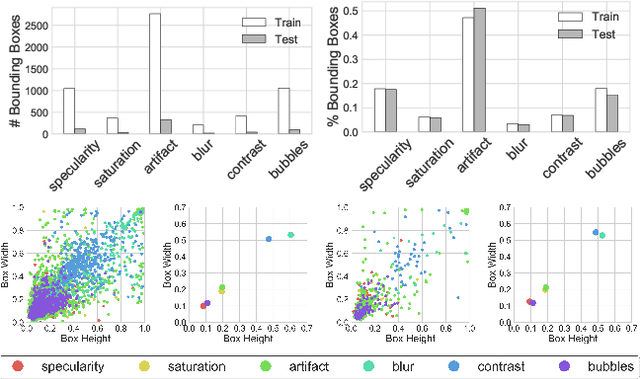

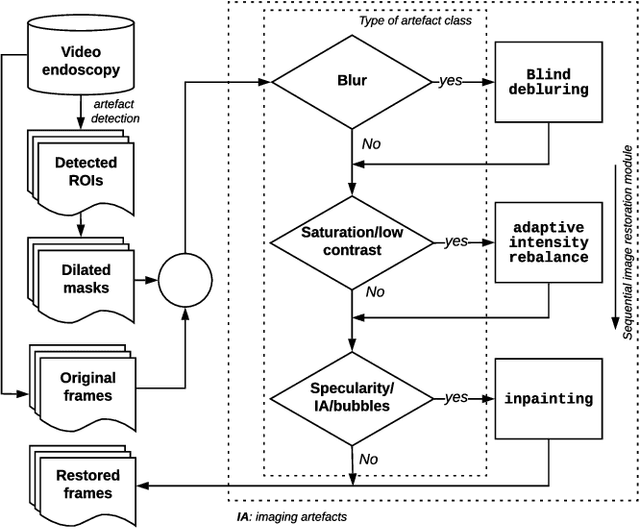

Abstract:Endoscopy is a routine imaging technique used for both diagnosis and minimally invasive surgical treatment. Artifacts such as motion blur, bubbles, specular reflections, floating objects and pixel saturation impede the visual interpretation and the automated analysis of endoscopy videos. Given the widespread use of endoscopy in different clinical applications, we contend that the robust and reliable identification of such artifacts and the automated restoration of corrupted video frames is a fundamental medical imaging problem. Existing state-of-the-art methods only deal with the detection and restoration of selected artifacts. However, typically endoscopy videos contain numerous artifacts which motivates to establish a comprehensive solution. We propose a fully automatic framework that can: 1) detect and classify six different primary artifacts, 2) provide a quality score for each frame and 3) restore mildly corrupted frames. To detect different artifacts our framework exploits fast multi-scale, single stage convolutional neural network detector. We introduce a quality metric to assess frame quality and predict image restoration success. Generative adversarial networks with carefully chosen regularization are finally used to restore corrupted frames. Our detector yields the highest mean average precision (mAP at 5% threshold) of 49.0 and the lowest computational time of 88 ms allowing for accurate real-time processing. Our restoration models for blind deblurring, saturation correction and inpainting demonstrate significant improvements over previous methods. On a set of 10 test videos we show that our approach preserves an average of 68.7% which is 25% more frames than that retained from the raw videos.
Improving Whole Slide Segmentation Through Visual Context - A Systematic Study
Jun 11, 2018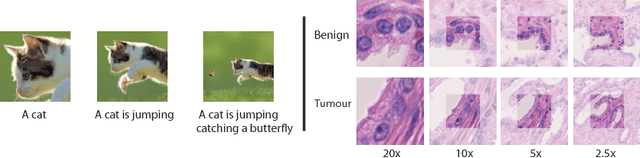
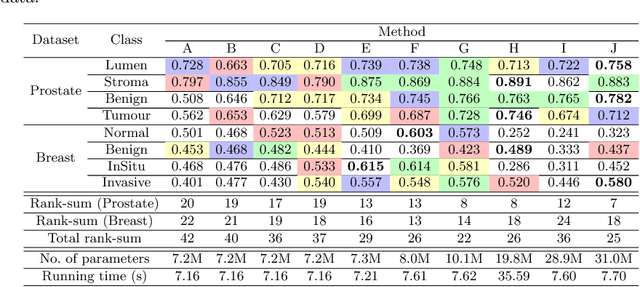
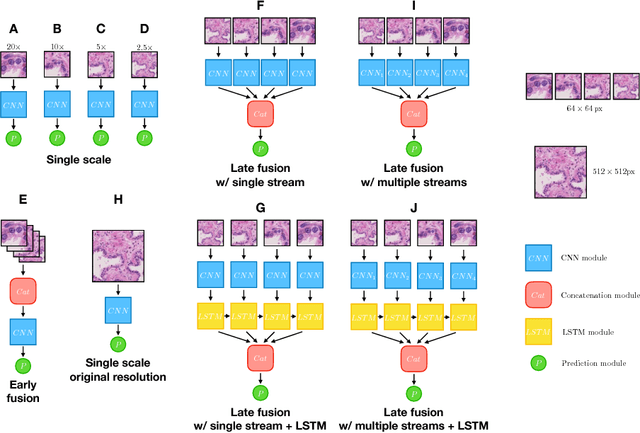
Abstract:While challenging, the dense segmentation of histology images is a necessary first step to assess changes in tissue architecture and cellular morphology. Although specific convolutional neural network architectures have been applied with great success to the problem, few effectively incorporate visual context information from multiple scales. With this paper, we present a systematic comparison of different architectures to assess how including multi-scale information affects segmentation performance. A publicly available breast cancer and a locally collected prostate cancer datasets are being utilised for this study. The results support our hypothesis that visual context and scale play a crucial role in histology image classification problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge