Jean-Christophe Lombardo
Unlocking Zero-Shot Plant Segmentation with Pl@ntNet Intelligence
Oct 14, 2025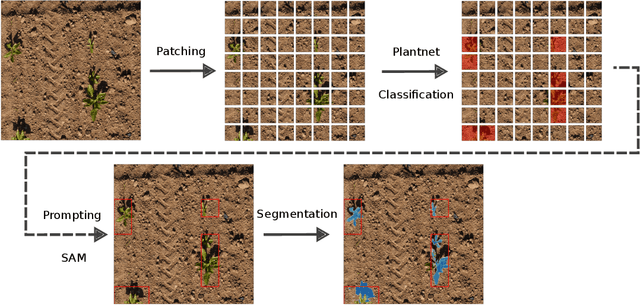
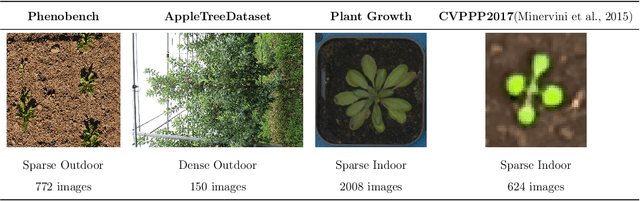
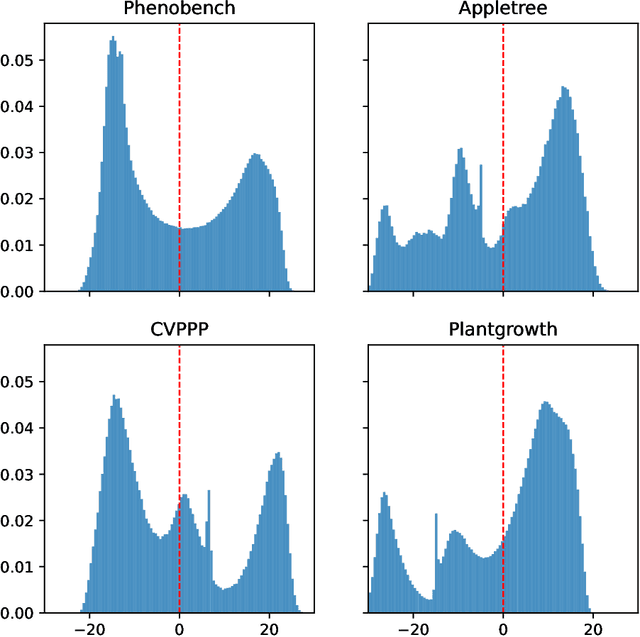
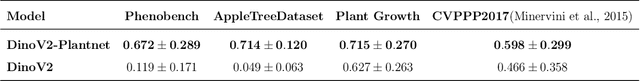
Abstract:We present a zero-shot segmentation approach for agricultural imagery that leverages Plantnet, a large-scale plant classification model, in conjunction with its DinoV2 backbone and the Segment Anything Model (SAM). Rather than collecting and annotating new datasets, our method exploits Plantnet's specialized plant representations to identify plant regions and produce coarse segmentation masks. These masks are then refined by SAM to yield detailed segmentations. We evaluate on four publicly available datasets of various complexity in terms of contrast including some where the limited size of the training data and complex field conditions often hinder purely supervised methods. Our results show consistent performance gains when using Plantnet-fine-tuned DinoV2 over the base DinoV2 model, as measured by the Jaccard Index (IoU). These findings highlight the potential of combining foundation models with specialized plant-centric models to alleviate the annotation bottleneck and enable effective segmentation in diverse agricultural scenarios.
Cooperative learning of Pl@ntNet's Artificial Intelligence algorithm: how does it work and how can we improve it?
Jun 05, 2024Abstract:Deep learning models for plant species identification rely on large annotated datasets. The PlantNet system enables global data collection by allowing users to upload and annotate plant observations, leading to noisy labels due to diverse user skills. Achieving consensus is crucial for training, but the vast scale of collected data makes traditional label aggregation strategies challenging. Existing methods either retain all observations, resulting in noisy training data or selectively keep those with sufficient votes, discarding valuable information. Additionally, as many species are rarely observed, user expertise can not be evaluated as an inter-user agreement: otherwise, botanical experts would have a lower weight in the AI training step than the average user. Our proposed label aggregation strategy aims to cooperatively train plant identification AI models. This strategy estimates user expertise as a trust score per user based on their ability to identify plant species from crowdsourced data. The trust score is recursively estimated from correctly identified species given the current estimated labels. This interpretable score exploits botanical experts' knowledge and the heterogeneity of users. Subsequently, our strategy removes unreliable observations but retains those with limited trusted annotations, unlike other approaches. We evaluate PlantNet's strategy on a released large subset of the PlantNet database focused on European flora, comprising over 6M observations and 800K users. We demonstrate that estimating users' skills based on the diversity of their expertise enhances labeling performance. Our findings emphasize the synergy of human annotation and data filtering in improving AI performance for a refined dataset. We explore incorporating AI-based votes alongside human input. This can further enhance human-AI interactions to detect unreliable observations.
Reproducible Performance Optimization of Complex Applications on the Edge-to-Cloud Continuum
Aug 04, 2021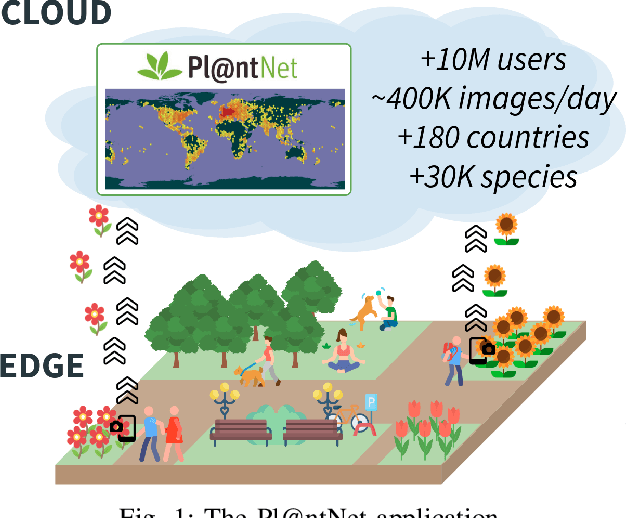
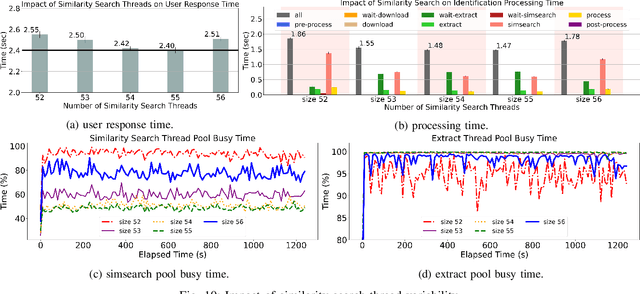
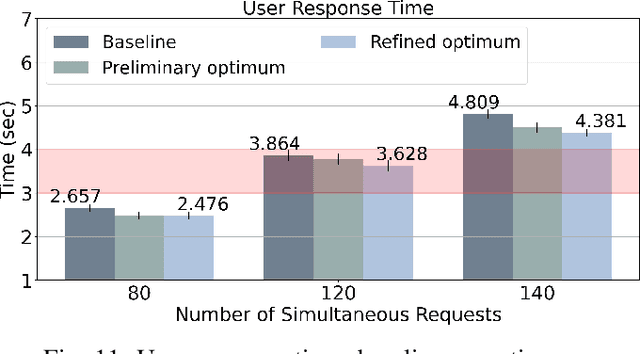
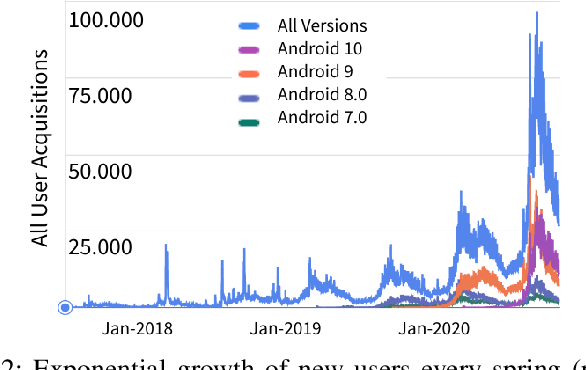
Abstract:In more and more application areas, we are witnessing the emergence of complex workflows that combine computing, analytics and learning. They often require a hybrid execution infrastructure with IoT devices interconnected to cloud/HPC systems (aka Computing Continuum). Such workflows are subject to complex constraints and requirements in terms of performance, resource usage, energy consumption and financial costs. This makes it challenging to optimize their configuration and deployment. We propose a methodology to support the optimization of real-life applications on the Edge-to-Cloud Continuum. We implement it as an extension of E2Clab, a previously proposed framework supporting the complete experimental cycle across the Edge-to-Cloud Continuum. Our approach relies on a rigorous analysis of possible configurations in a controlled testbed environment to understand their behaviour and related performance trade-offs. We illustrate our methodology by optimizing Pl@ntNet, a world-wide plant identification application. Our methodology can be generalized to other applications in the Edge-to-Cloud Continuum.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge