Alexis Joly
ZENITH, LIRMM, UM
Audio-to-Image Bird Species Retrieval without Audio-Image Pairs via Text Distillation
Jan 31, 2026Abstract:Audio-to-image retrieval offers an interpretable alternative to audio-only classification for bioacoustic species recognition, but learning aligned audio-image representations is challenging due to the scarcity of paired audio-image data. We propose a simple and data-efficient approach that enables audio-to-image retrieval without any audio-image supervision. Our proposed method uses text as a semantic intermediary: we distill the text embedding space of a pretrained image-text model (BioCLIP-2), which encodes rich visual and taxonomic structure, into a pretrained audio-text model (BioLingual) by fine-tuning its audio encoder with a contrastive objective. This distillation transfers visually grounded semantics into the audio representation, inducing emergent alignment between audio and image embeddings without using images during training. We evaluate the resulting model on multiple bioacoustic benchmarks. The distilled audio encoder preserves audio discriminative power while substantially improving audio-text alignment on focal recordings and soundscape datasets. Most importantly, on the SSW60 benchmark, the proposed approach achieves strong audio-to-image retrieval performance exceeding baselines based on zero-shot model combinations or learned mappings between text embeddings, despite not training on paired audio-image data. These results demonstrate that indirect semantic transfer through text is sufficient to induce meaningful audio-image alignment, providing a practical solution for visually grounded species recognition in data-scarce bioacoustic settings.
Compact Hypercube Embeddings for Fast Text-based Wildlife Observation Retrieval
Jan 30, 2026Abstract:Large-scale biodiversity monitoring platforms increasingly rely on multimodal wildlife observations. While recent foundation models enable rich semantic representations across vision, audio, and language, retrieving relevant observations from massive archives remains challenging due to the computational cost of high-dimensional similarity search. In this work, we introduce compact hypercube embeddings for fast text-based wildlife observation retrieval, a framework that enables efficient text-based search over large-scale wildlife image and audio databases using compact binary representations. Building on the cross-view code alignment hashing framework, we extend lightweight hashing beyond a single-modality setup to align natural language descriptions with visual or acoustic observations in a shared Hamming space. Our approach leverages pretrained wildlife foundation models, including BioCLIP and BioLingual, and adapts them efficiently for hashing using parameter-efficient fine-tuning. We evaluate our method on large-scale benchmarks, including iNaturalist2024 for text-to-image retrieval and iNatSounds2024 for text-to-audio retrieval, as well as multiple soundscape datasets to assess robustness under domain shift. Results show that retrieval using discrete hypercube embeddings achieves competitive, and in several cases superior, performance compared to continuous embeddings, while drastically reducing memory and search cost. Moreover, we observe that the hashing objective consistently improves the underlying encoder representations, leading to stronger retrieval and zero-shot generalization. These results demonstrate that binary, language-based retrieval enables scalable and efficient search over large wildlife archives for biodiversity monitoring systems.
GeoPl@ntNet: A Platform for Exploring Essential Biodiversity Variables
Nov 16, 2025Abstract:This paper describes GeoPl@ntNet, an interactive web application designed to make Essential Biodiversity Variables accessible and understandable to everyone through dynamic maps and fact sheets. Its core purpose is to allow users to explore high-resolution AI-generated maps of species distributions, habitat types, and biodiversity indicators across Europe. These maps, developed through a cascading pipeline involving convolutional neural networks and large language models, provide an intuitive yet information-rich interface to better understand biodiversity, with resolutions as precise as 50x50 meters. The website also enables exploration of specific regions, allowing users to select areas of interest on the map (e.g., urban green spaces, protected areas, or riverbanks) to view local species and their coverage. Additionally, GeoPl@ntNet generates comprehensive reports for selected regions, including insights into the number of protected species, invasive species, and endemic species.
Image Hashing via Cross-View Code Alignment in the Age of Foundation Models
Nov 03, 2025Abstract:Efficient large-scale retrieval requires representations that are both compact and discriminative. Foundation models provide powerful visual and multimodal embeddings, but nearest neighbor search in these high-dimensional spaces is computationally expensive. Hashing offers an efficient alternative by enabling fast Hamming distance search with binary codes, yet existing approaches often rely on complex pipelines, multi-term objectives, designs specialized for a single learning paradigm, and long training times. We introduce CroVCA (Cross-View Code Alignment), a simple and unified principle for learning binary codes that remain consistent across semantically aligned views. A single binary cross-entropy loss enforces alignment, while coding-rate maximization serves as an anti-collapse regularizer to promote balanced and diverse codes. To implement this, we design HashCoder, a lightweight MLP hashing network with a final batch normalization layer to enforce balanced codes. HashCoder can be used as a probing head on frozen embeddings or to adapt encoders efficiently via LoRA fine-tuning. Across benchmarks, CroVCA achieves state-of-the-art results in just 5 training epochs. At 16 bits, it particularly well-for instance, unsupervised hashing on COCO completes in under 2 minutes and supervised hashing on ImageNet100 in about 3 minutes on a single GPU. These results highlight CroVCA's efficiency, adaptability, and broad applicability.
Unlocking Zero-Shot Plant Segmentation with Pl@ntNet Intelligence
Oct 14, 2025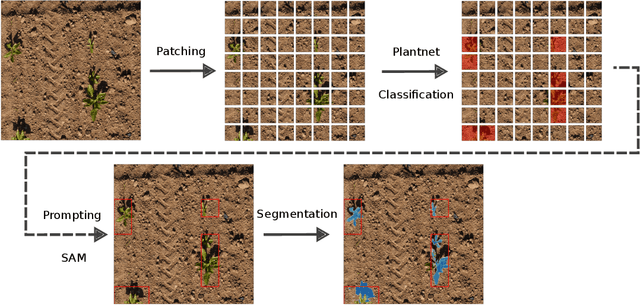
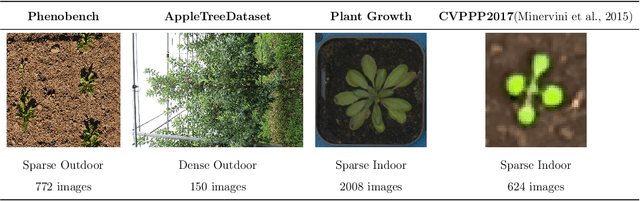
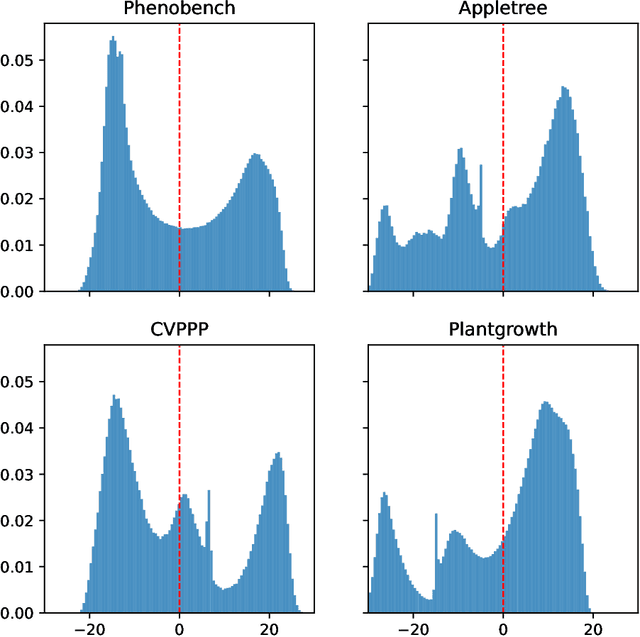
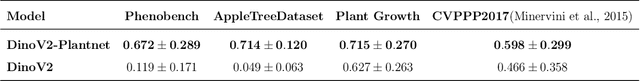
Abstract:We present a zero-shot segmentation approach for agricultural imagery that leverages Plantnet, a large-scale plant classification model, in conjunction with its DinoV2 backbone and the Segment Anything Model (SAM). Rather than collecting and annotating new datasets, our method exploits Plantnet's specialized plant representations to identify plant regions and produce coarse segmentation masks. These masks are then refined by SAM to yield detailed segmentations. We evaluate on four publicly available datasets of various complexity in terms of contrast including some where the limited size of the training data and complex field conditions often hinder purely supervised methods. Our results show consistent performance gains when using Plantnet-fine-tuned DinoV2 over the base DinoV2 model, as measured by the Jaccard Index (IoU). These findings highlight the potential of combining foundation models with specialized plant-centric models to alleviate the annotation bottleneck and enable effective segmentation in diverse agricultural scenarios.
Overview of PlantCLEF 2024: multi-species plant identification in vegetation plot images
Sep 19, 2025



Abstract:Plot images are essential for ecological studies, enabling standardized sampling, biodiversity assessment, long-term monitoring and remote, large-scale surveys. Plot images are typically fifty centimetres or one square meter in size, and botanists meticulously identify all the species found there. The integration of AI could significantly improve the efficiency of specialists, helping them to extend the scope and coverage of ecological studies. To evaluate advances in this regard, the PlantCLEF 2024 challenge leverages a new test set of thousands of multi-label images annotated by experts and covering over 800 species. In addition, it provides a large training set of 1.7 million individual plant images as well as state-of-the-art vision transformer models pre-trained on this data. The task is evaluated as a (weakly-labeled) multi-label classification task where the aim is to predict all the plant species present on a high-resolution plot image (using the single-label training data). In this paper, we provide an detailed description of the data, the evaluation methodology, the methods and models employed by the participants and the results achieved.
Hashing-Baseline: Rethinking Hashing in the Age of Pretrained Models
Sep 17, 2025



Abstract:Information retrieval with compact binary embeddings, also referred to as hashing, is crucial for scalable fast search applications, yet state-of-the-art hashing methods require expensive, scenario-specific training. In this work, we introduce Hashing-Baseline, a strong training-free hashing method leveraging powerful pretrained encoders that produce rich pretrained embeddings. We revisit classical, training-free hashing techniques: principal component analysis, random orthogonal projection, and threshold binarization, to produce a strong baseline for hashing. Our approach combines these techniques with frozen embeddings from state-of-the-art vision and audio encoders to yield competitive retrieval performance without any additional learning or fine-tuning. To demonstrate the generality and effectiveness of this approach, we evaluate it on standard image retrieval benchmarks as well as a newly introduced benchmark for audio hashing.
The point is the mask: scaling coral reef segmentation with weak supervision
Aug 26, 2025Abstract:Monitoring coral reefs at large spatial scales remains an open challenge, essential for assessing ecosystem health and informing conservation efforts. While drone-based aerial imagery offers broad spatial coverage, its limited resolution makes it difficult to reliably distinguish fine-scale classes, such as coral morphotypes. At the same time, obtaining pixel-level annotations over large spatial extents is costly and labor-intensive, limiting the scalability of deep learning-based segmentation methods for aerial imagery. We present a multi-scale weakly supervised semantic segmentation framework that addresses this challenge by transferring fine-scale ecological information from underwater imagery to aerial data. Our method enables large-scale coral reef mapping from drone imagery with minimal manual annotation, combining classification-based supervision, spatial interpolation and self-distillation techniques. We demonstrate the efficacy of the approach, enabling large-area segmentation of coral morphotypes and demonstrating flexibility for integrating new classes. This study presents a scalable, cost-effective methodology for high-resolution reef monitoring, combining low-cost data collection, weakly supervised deep learning and multi-scale remote sensing.
Can Masked Autoencoders Also Listen to Birds?
Apr 17, 2025Abstract:Masked Autoencoders (MAEs) pretrained on AudioSet fail to capture the fine-grained acoustic characteristics of specialized domains such as bioacoustic monitoring. Bird sound classification is critical for assessing environmental health, yet general-purpose models inadequately address its unique acoustic challenges. To address this, we introduce Bird-MAE, a domain-specialized MAE pretrained on the large-scale BirdSet dataset. We explore adjustments to pretraining, fine-tuning and utilizing frozen representations. Bird-MAE achieves state-of-the-art results across all BirdSet downstream tasks, substantially improving multi-label classification performance compared to the general-purpose Audio-MAE baseline. Additionally, we propose prototypical probing, a parameter-efficient method for leveraging MAEs' frozen representations. Bird-MAE's prototypical probes outperform linear probing by up to 37\% in MAP and narrow the gap to fine-tuning to approximately 3\% on average on BirdSet.
Mapping biodiversity at very-high resolution in Europe
Apr 07, 2025



Abstract:This paper describes a cascading multimodal pipeline for high-resolution biodiversity mapping across Europe, integrating species distribution modeling, biodiversity indicators, and habitat classification. The proposed pipeline first predicts species compositions using a deep-SDM, a multimodal model trained on remote sensing, climate time series, and species occurrence data at 50x50m resolution. These predictions are then used to generate biodiversity indicator maps and classify habitats with Pl@ntBERT, a transformer-based LLM designed for species-to-habitat mapping. With this approach, continental-scale species distribution maps, biodiversity indicator maps, and habitat maps are produced, providing fine-grained ecological insights. Unlike traditional methods, this framework enables joint modeling of interspecies dependencies, bias-aware training with heterogeneous presence-absence data, and large-scale inference from multi-source remote sensing inputs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge