Jama Hussein Mohamud
Leveraging LLMs as Meta-Judges: A Multi-Agent Framework for Evaluating LLM Judgments
Apr 23, 2025Abstract:Large language models (LLMs) are being widely applied across various fields, but as tasks become more complex, evaluating their responses is increasingly challenging. Compared to human evaluators, the use of LLMs to support performance evaluation offers a more efficient alternative. However, most studies focus mainly on aligning LLMs' judgments with human preferences, overlooking the existence of biases and mistakes in human judgment. Furthermore, how to select suitable LLM judgments given multiple potential LLM responses remains underexplored. To address these two aforementioned issues, we propose a three-stage meta-judge selection pipeline: 1) developing a comprehensive rubric with GPT-4 and human experts, 2) using three advanced LLM agents to score judgments, and 3) applying a threshold to filter out low-scoring judgments. Compared to methods using a single LLM as both judge and meta-judge, our pipeline introduces multi-agent collaboration and a more comprehensive rubric. Experimental results on the JudgeBench dataset show about 15.55\% improvement compared to raw judgments and about 8.37\% improvement over the single-agent baseline. Our work demonstrates the potential of LLMs as meta-judges and lays the foundation for future research on constructing preference datasets for LLM-as-a-judge reinforcement learning.
Towards Foundational Models for Molecular Learning on Large-Scale Multi-Task Datasets
Oct 18, 2023



Abstract:Recently, pre-trained foundation models have enabled significant advancements in multiple fields. In molecular machine learning, however, where datasets are often hand-curated, and hence typically small, the lack of datasets with labeled features, and codebases to manage those datasets, has hindered the development of foundation models. In this work, we present seven novel datasets categorized by size into three distinct categories: ToyMix, LargeMix and UltraLarge. These datasets push the boundaries in both the scale and the diversity of supervised labels for molecular learning. They cover nearly 100 million molecules and over 3000 sparsely defined tasks, totaling more than 13 billion individual labels of both quantum and biological nature. In comparison, our datasets contain 300 times more data points than the widely used OGB-LSC PCQM4Mv2 dataset, and 13 times more than the quantum-only QM1B dataset. In addition, to support the development of foundational models based on our proposed datasets, we present the Graphium graph machine learning library which simplifies the process of building and training molecular machine learning models for multi-task and multi-level molecular datasets. Finally, we present a range of baseline results as a starting point of multi-task and multi-level training on these datasets. Empirically, we observe that performance on low-resource biological datasets show improvement by also training on large amounts of quantum data. This indicates that there may be potential in multi-task and multi-level training of a foundation model and fine-tuning it to resource-constrained downstream tasks.
Fast Development of ASR in African Languages using Self Supervised Speech Representation Learning
Mar 16, 2021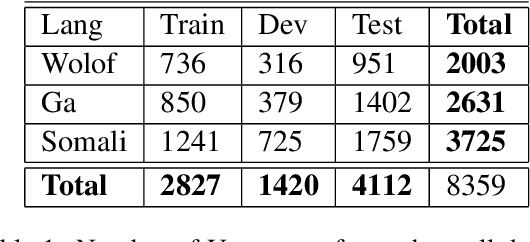
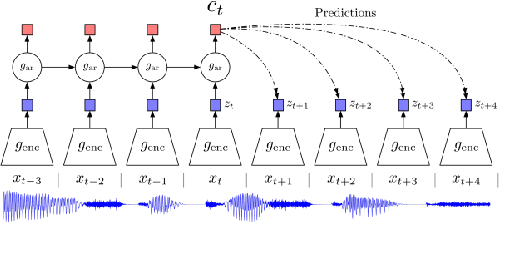
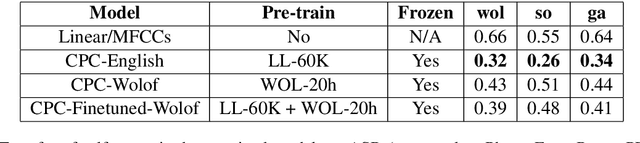
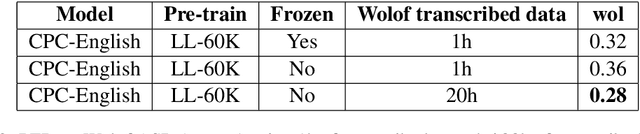
Abstract:This paper describes the results of an informal collaboration launched during the African Master of Machine Intelligence (AMMI) in June 2020. After a series of lectures and labs on speech data collection using mobile applications and on self-supervised representation learning from speech, a small group of students and the lecturer continued working on automatic speech recognition (ASR) project for three languages: Wolof, Ga, and Somali. This paper describes how data was collected and ASR systems developed with a small amount (1h) of transcribed speech as training data. In these low resource conditions, pre-training a model on large amounts of raw speech was fundamental for the efficiency of ASR systems developed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge