Isabella Cama
Probabilistic approach to longitudinal response prediction: application to radiomics from brain cancer imaging
May 12, 2025Abstract:Longitudinal imaging analysis tracks disease progression and treatment response over time, providing dynamic insights into treatment efficacy and disease evolution. Radiomic features extracted from medical imaging can support the study of disease progression and facilitate longitudinal prediction of clinical outcomes. This study presents a probabilistic model for longitudinal response prediction, integrating baseline features with intermediate follow-ups. The probabilistic nature of the model naturally allows to handle the instrinsic uncertainty of the longitudinal prediction of disease progression. We evaluate the proposed model against state-of-the-art disease progression models in both a synthetic scenario and using a brain cancer dataset. Results demonstrate that the approach is competitive against existing methods while uniquely accounting for uncertainty and controlling the growth of problem dimensionality, eliminating the need for data from intermediate follow-ups.
DISARM++: Beyond scanner-free harmonization
May 06, 2025



Abstract:Harmonization of T1-weighted MR images across different scanners is crucial for ensuring consistency in neuroimaging studies. This study introduces a novel approach to direct image harmonization, moving beyond feature standardization to ensure that extracted features remain inherently reliable for downstream analysis. Our method enables image transfer in two ways: (1) mapping images to a scanner-free space for uniform appearance across all scanners, and (2) transforming images into the domain of a specific scanner used in model training, embedding its unique characteristics. Our approach presents strong generalization capability, even for unseen scanners not included in the training phase. We validated our method using MR images from diverse cohorts, including healthy controls, traveling subjects, and individuals with Alzheimer's disease (AD). The model's effectiveness is tested in multiple applications, such as brain age prediction (R2 = 0.60 \pm 0.05), biomarker extraction, AD classification (Test Accuracy = 0.86 \pm 0.03), and diagnosis prediction (AUC = 0.95). In all cases, our harmonization technique outperforms state-of-the-art methods, showing improvements in both reliability and predictive accuracy. Moreover, our approach eliminates the need for extensive preprocessing steps, such as skull-stripping, which can introduce errors by misclassifying brain and non-brain structures. This makes our method particularly suitable for applications that require full-head analysis, including research on head trauma and cranial deformities. Additionally, our harmonization model does not require retraining for new datasets, allowing smooth integration into various neuroimaging workflows. By ensuring scanner-invariant image quality, our approach provides a robust and efficient solution for improving neuroimaging studies across diverse settings. The code is available at this link.
Segmentation variability and radiomics stability for predicting Triple-Negative Breast Cancer subtype using Magnetic Resonance Imaging
Apr 02, 2025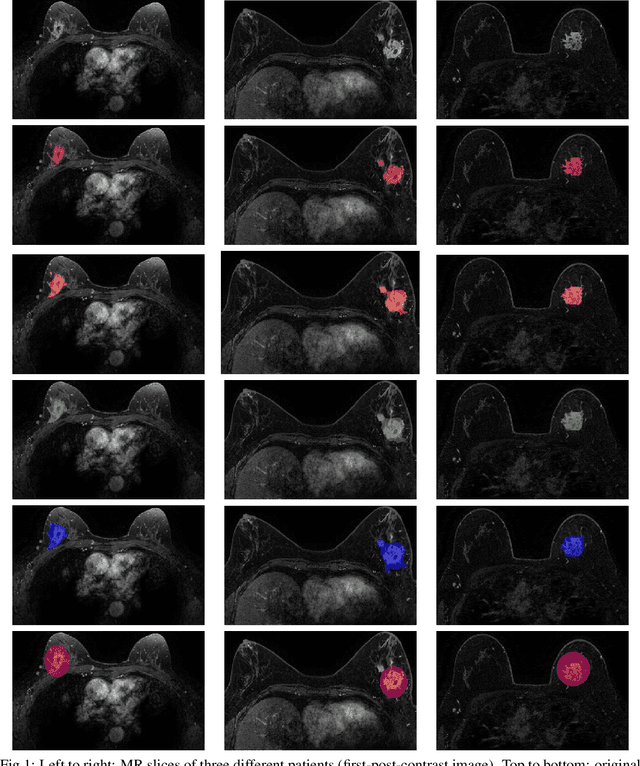
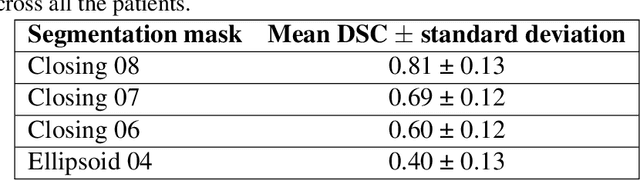
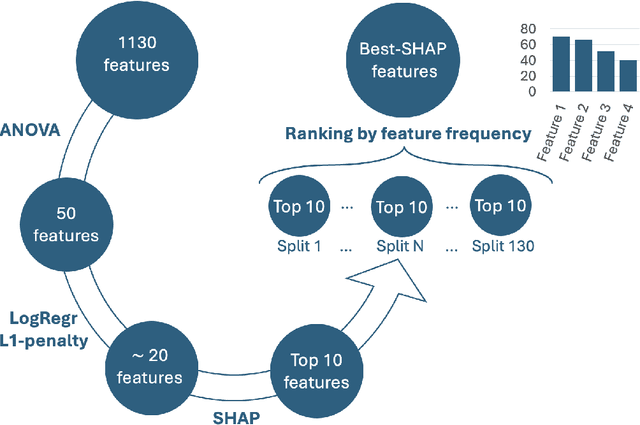
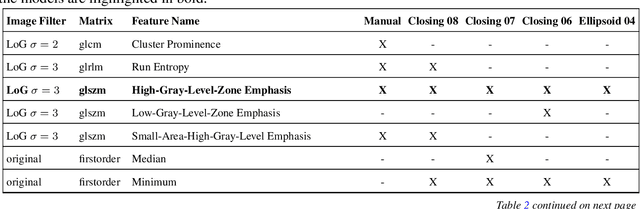
Abstract:Most papers caution against using predictive models for disease stratification based on unselected radiomic features, as these features are affected by contouring variability. Instead, they advocate for the use of the Intraclass Correlation Coefficient (ICC) as a measure of stability for feature selection. However, the direct effect of segmentation variability on the predictive models is rarely studied. This study investigates the impact of segmentation variability on feature stability and predictive performance in radiomics-based prediction of Triple-Negative Breast Cancer (TNBC) subtype using Magnetic Resonance Imaging. A total of 244 images from the Duke dataset were used, with segmentation variability introduced through modifications of manual segmentations. For each mask, explainable radiomic features were selected using the Shapley Additive exPlanations method and used to train logistic regression models. Feature stability across segmentations was assessed via ICC, Pearson's correlation, and reliability scores quantifying the relationship between feature stability and segmentation variability. Results indicate that segmentation accuracy does not significantly impact predictive performance. While incorporating peritumoral information may reduce feature reproducibility, it does not diminish feature predictive capability. Moreover, feature selection in predictive models is not inherently tied to feature stability with respect to segmentation, suggesting that an overreliance on ICC or reliability scores for feature selection might exclude valuable predictive features.
Three-dimensional numerical schemes for the segmentation of the psoas muscle in X-ray computed tomography images
Dec 10, 2023Abstract:The analysis of the psoas muscle in morphological and functional imaging has proved to be an accurate approach to assess sarcopenia, i.e. a systemic loss of skeletal muscle mass and function that may be correlated to multifactorial etiological aspects. The inclusion of sarcopenia assessment into a radiological workflow would need the implementation of computational pipelines for image processing that guarantee segmentation reliability and a significant degree of automation. The present study utilizes three-dimensional numerical schemes for psoas segmentation in low-dose X-ray computed tomography images. Specifically, here we focused on the level set methodology and compared the performances of two standard approaches, a classical evolution model and a three-dimension geodesic model, with the performances of an original first-order modification of this latter one. The results of this analysis show that these gradient-based schemes guarantee reliability with respect to manual segmentation and that the first-order scheme requires a computational burden that is significantly smaller than the one needed by the second-order approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge