Hongjun Suh
Frequency Composition for Compressed and Domain-Adaptive Neural Networks
May 27, 2025Abstract:Modern on-device neural network applications must operate under resource constraints while adapting to unpredictable domain shifts. However, this combined challenge-model compression and domain adaptation-remains largely unaddressed, as prior work has tackled each issue in isolation: compressed networks prioritize efficiency within a fixed domain, whereas large, capable models focus on handling domain shifts. In this work, we propose CoDA, a frequency composition-based framework that unifies compression and domain adaptation. During training, CoDA employs quantization-aware training (QAT) with low-frequency components, enabling a compressed model to selectively learn robust, generalizable features. At test time, it refines the compact model in a source-free manner (i.e., test-time adaptation, TTA), leveraging the full-frequency information from incoming data to adapt to target domains while treating high-frequency components as domain-specific cues. LFC are aligned with the trained distribution, while HFC unique to the target distribution are solely utilized for batch normalization. CoDA can be integrated synergistically into existing QAT and TTA methods. CoDA is evaluated on widely used domain-shift benchmarks, including CIFAR10-C and ImageNet-C, across various model architectures. With significant compression, it achieves accuracy improvements of 7.96%p on CIFAR10-C and 5.37%p on ImageNet-C over the full-precision TTA baseline.
Non-Volatile Memory Accelerated Geometric Multi-Scale Resolution Analysis
Feb 21, 2022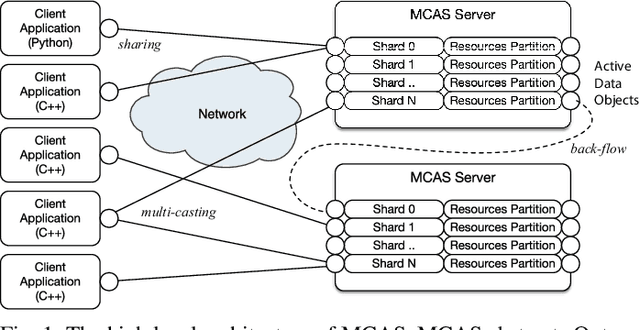
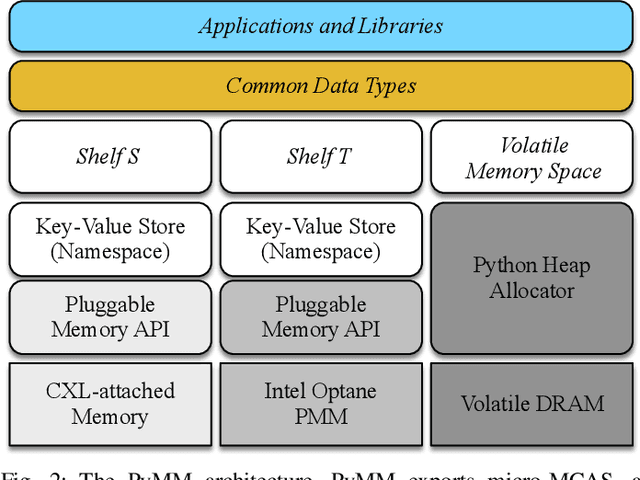
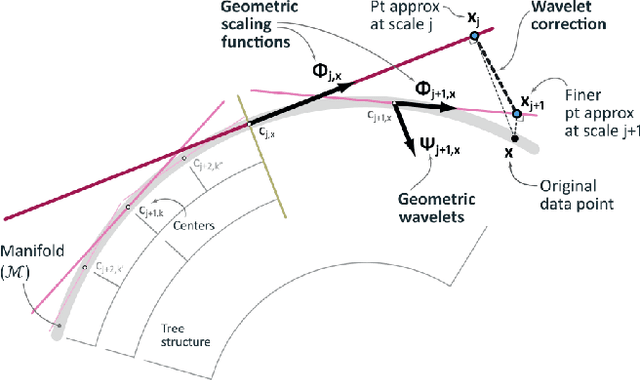
Abstract:Dimensionality reduction algorithms are standard tools in a researcher's toolbox. Dimensionality reduction algorithms are frequently used to augment downstream tasks such as machine learning, data science, and also are exploratory methods for understanding complex phenomena. For instance, dimensionality reduction is commonly used in Biology as well as Neuroscience to understand data collected from biological subjects. However, dimensionality reduction techniques are limited by the von-Neumann architectures that they execute on. Specifically, data intensive algorithms such as dimensionality reduction techniques often require fast, high capacity, persistent memory which historically hardware has been unable to provide at the same time. In this paper, we present a re-implementation of an existing dimensionality reduction technique called Geometric Multi-Scale Resolution Analysis (GMRA) which has been accelerated via novel persistent memory technology called Memory Centric Active Storage (MCAS). Our implementation uses a specialized version of MCAS called PyMM that provides native support for Python datatypes including NumPy arrays and PyTorch tensors. We compare our PyMM implementation against a DRAM implementation, and show that when data fits in DRAM, PyMM offers competitive runtimes. When data does not fit in DRAM, our PyMM implementation is still able to process the data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge