Hemant D. Tagare
VidFuncta: Towards Generalizable Neural Representations for Ultrasound Videos
Jul 29, 2025Abstract:Ultrasound is widely used in clinical care, yet standard deep learning methods often struggle with full video analysis due to non-standardized acquisition and operator bias. We offer a new perspective on ultrasound video analysis through implicit neural representations (INRs). We build on Functa, an INR framework in which each image is represented by a modulation vector that conditions a shared neural network. However, its extension to the temporal domain of medical videos remains unexplored. To address this gap, we propose VidFuncta, a novel framework that leverages Functa to encode variable-length ultrasound videos into compact, time-resolved representations. VidFuncta disentangles each video into a static video-specific vector and a sequence of time-dependent modulation vectors, capturing both temporal dynamics and dataset-level redundancies. Our method outperforms 2D and 3D baselines on video reconstruction and enables downstream tasks to directly operate on the learned 1D modulation vectors. We validate VidFuncta on three public ultrasound video datasets -- cardiac, lung, and breast -- and evaluate its downstream performance on ejection fraction prediction, B-line detection, and breast lesion classification. These results highlight the potential of VidFuncta as a generalizable and efficient representation framework for ultrasound videos. Our code is publicly available under https://github.com/JuliaWolleb/VidFuncta_public.
Self-normalized Classification of Parkinson's Disease DaTscan Images
Dec 27, 2021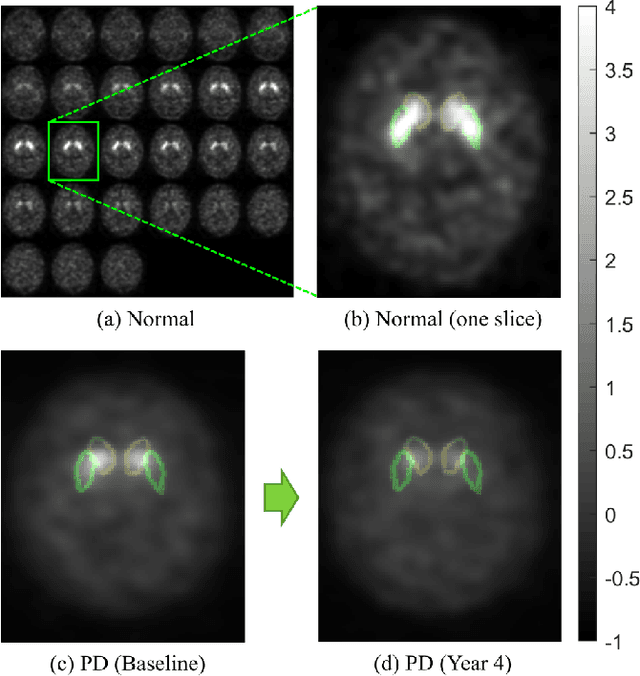
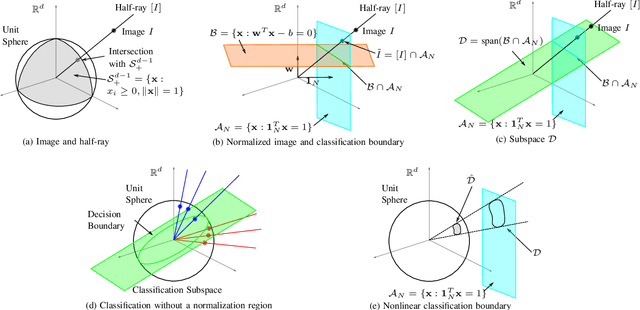
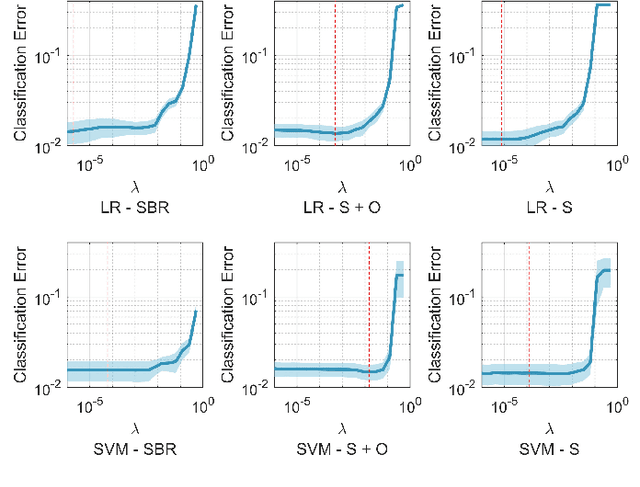
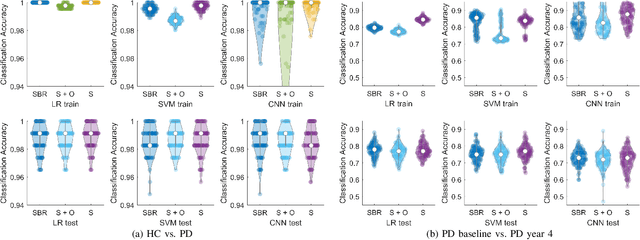
Abstract:Classifying SPECT images requires a preprocessing step which normalizes the images using a normalization region. The choice of the normalization region is not standard, and using different normalization regions introduces normalization region-dependent variability. This paper mathematically analyzes the effect of the normalization region to show that normalized-classification is exactly equivalent to a subspace separation of the half rays of the images under multiplicative equivalence. Using this geometry, a new self-normalized classification strategy is proposed. This strategy eliminates the normalizing region altogether. The theory is used to classify DaTscan images of 365 Parkinson's disease (PD) subjects and 208 healthy control (HC) subjects from the Parkinson's Progression Marker Initiative (PPMI). The theory is also used to understand PD progression from baseline to year 4.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge