Harald Becher
Accelerated 3D-3D rigid registration of echocardiographic images obtained from apical window using particle filter
Apr 28, 2025



Abstract:The perfect alignment of 3D echocardiographic images captured from various angles has improved image quality and broadened the field of view. This study proposes an accelerated sequential Monte Carlo (SMC) algorithm for 3D-3D rigid registration of transthoracic echocardiographic images with significant and limited overlap taken from apical window that is robust to the noise and intensity variation in ultrasound images. The algorithm estimates the translational and rotational components of the rigid transform through an iterative process and requires an initial approximation of the rotation and translation limits. We perform registration in two ways: the image-based registration computes the transform to align the end-diastolic frame of the apical nonstandard image to the apical standard image and applies the same transform to all frames of the cardiac cycle, whereas the mask-based registration approach uses the binary masks of the left ventricle in the same way. The SMC and exhaustive search (EX) algorithms were evaluated for 4D temporal sequences recorded from 7 volunteers who participated in a study conducted at the Mazankowski Alberta Heart Institute. The evaluations demonstrate that the mask-based approach of the accelerated SMC yielded a Dice score value of 0.819 +/- 0.045 for the left ventricle and gained 16.7x speedup compared to the CPU version of the SMC algorithm.
A New Semi-Automated Algorithm for Volumetric Segmentation of the Left Ventricle in Temporal 3D Echocardiography Sequences
Sep 03, 2021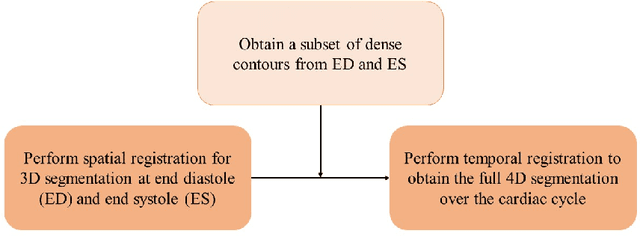
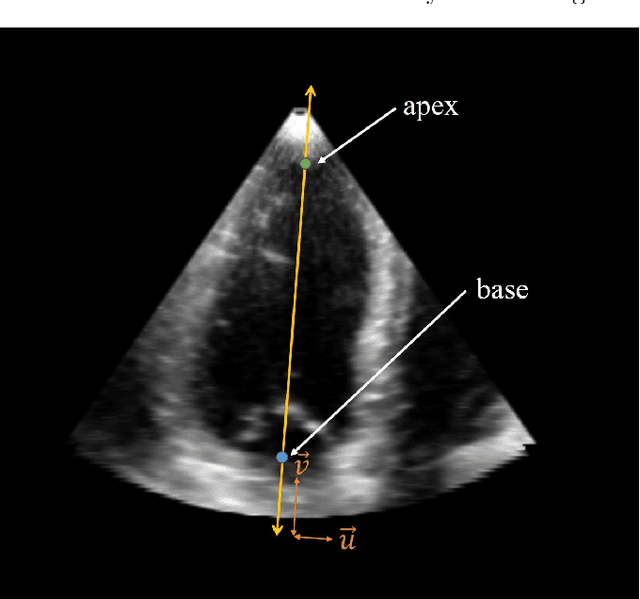
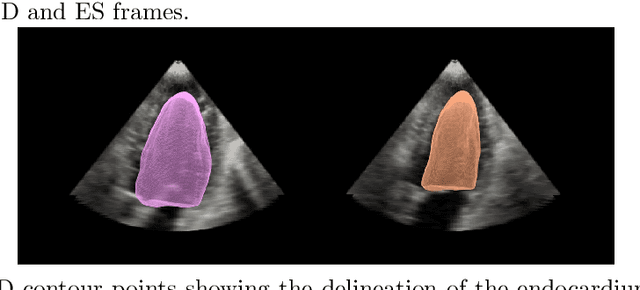
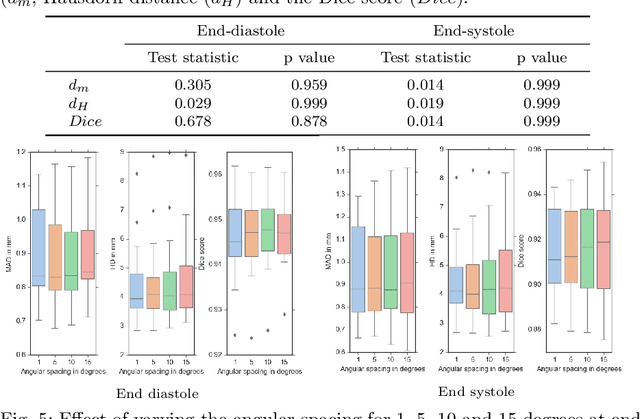
Abstract:Purpose: Echocardiography is commonly used as a non-invasive imaging tool in clinical practice for the assessment of cardiac function. However, delineation of the left ventricle is challenging due to the inherent properties of ultrasound imaging, such as the presence of speckle noise and the low signal-to-noise ratio. Methods: We propose a semi-automated segmentation algorithm for the delineation of the left ventricle in temporal 3D echocardiography sequences. The method requires minimal user interaction and relies on a diffeomorphic registration approach. Advantages of the method include no dependence on prior geometrical information, training data, or registration from an atlas. Results: The method was evaluated using three-dimensional ultrasound scan sequences from 18 patients from the Mazankowski Alberta Heart Institute, Edmonton, Canada, and compared to manual delineations provided by an expert cardiologist and four other registration algorithms. The segmentation approach yielded the following results over the cardiac cycle: a mean absolute difference of 1.01 (0.21) mm, a Hausdorff distance of 4.41 (1.43) mm, and a Dice overlap score of 0.93 (0.02). Conclusions: The method performed well compared to the four other registration algorithms.
* 22 pages, 8 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge