Haifa Alrdahi
Aspect-based Sentiment Evaluation of Chess Moves (ASSESS): an NLP-based Method for Evaluating Chess Strategies from Textbooks
May 10, 2024



Abstract:The chess domain is well-suited for creating an artificial intelligence (AI) system that mimics real-world challenges, including decision-making. Throughout the years, minimal attention has been paid to investigating insights derived from unstructured chess data sources. In this study, we examine the complicated relationships between multiple referenced moves in a chess-teaching textbook, and propose a novel method designed to encapsulate chess knowledge derived from move-action phrases. This study investigates the feasibility of using a modified sentiment analysis method as a means for evaluating chess moves based on text. Our proposed Aspect-Based Sentiment Analysis (ABSA) method represents an advancement in evaluating the sentiment associated with referenced chess moves. By extracting insights from move-action phrases, our approach aims to provide a more fine-grained and contextually aware `chess move'-based sentiment classification. Through empirical experiments and analysis, we evaluate the performance of our fine-tuned ABSA model, presenting results that confirm the efficiency of our approach in advancing aspect-based sentiment classification within the chess domain. This research contributes to the area of game-playing by machines and shows the practical applicability of leveraging NLP techniques to understand the context of strategic games.
Learning to Play Chess from Textbooks : a Corpus for Evaluating Chess Moves based on Sentiment Analysis
Oct 31, 2023Abstract:Learning chess strategies has been investigated widely, with most studies focussing on learning from previous games using search algorithms. Chess textbooks encapsulate grandmaster knowledge, explain playing strategies and require a smaller search space compared to traditional chess agents. This paper examines chess textbooks as a new knowledge source for enabling machines to learn how to play chess -- a resource that has not been explored previously. We developed the LEAP corpus, a first and new heterogeneous dataset with structured (chess move notations and board states) and unstructured data (textual descriptions) collected from a chess textbook containing 1164 sentences discussing strategic moves from 91 games. We firstly labelled the sentences based on their relevance, i.e., whether they are discussing a move. Each relevant sentence was then labelled according to its sentiment towards the described move. We performed empirical experiments that assess the performance of various transformer-based baseline models for sentiment analysis. Our results demonstrate the feasibility of employing transformer-based sentiment analysis models for evaluating chess moves, with the best performing model obtaining a weighted micro F_1 score of 68%. Finally, we synthesised the LEAP corpus to create a larger dataset, which can be used as a solution to the limited textual resource in the chess domain.
MedMine: Examining Pre-trained Language Models on Medication Mining
Aug 08, 2023



Abstract:Automatic medication mining from clinical and biomedical text has become a popular topic due to its real impact on healthcare applications and the recent development of powerful language models (LMs). However, fully-automatic extraction models still face obstacles to be overcome such that they can be deployed directly into clinical practice for better impacts. Such obstacles include their imbalanced performances on different entity types and clinical events. In this work, we examine current state-of-the-art pre-trained language models (PLMs) on such tasks, via fine-tuning including the monolingual model Med7 and multilingual large language model (LLM) XLM-RoBERTa. We compare their advantages and drawbacks using historical medication mining shared task data sets from n2c2-2018 challenges. We report the findings we get from these fine-tuning experiments such that they can facilitate future research on addressing them, for instance, how to combine their outputs, merge such models, or improve their overall accuracy by ensemble learning and data augmentation. MedMine is part of the M3 Initiative \url{https://github.com/HECTA-UoM/M3}
GNTeam at 2018 n2c2: Feature-augmented BiLSTM-CRF for drug-related entity recognition in hospital discharge summaries
Sep 23, 2019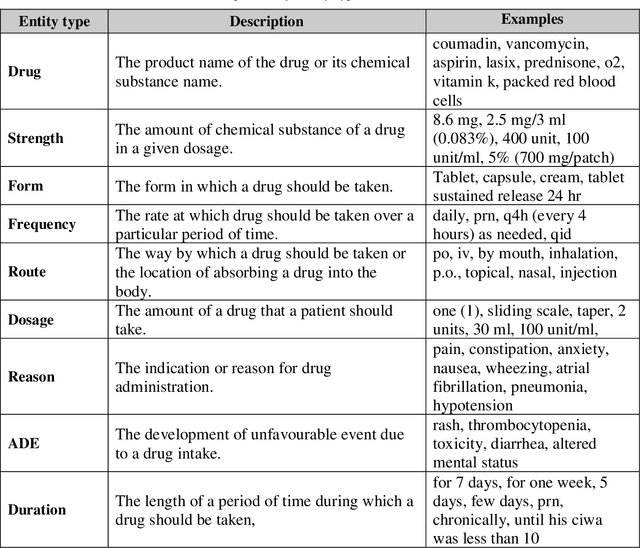
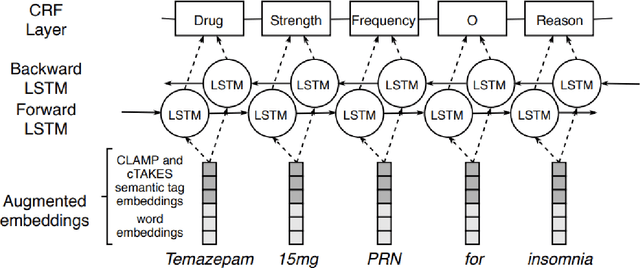
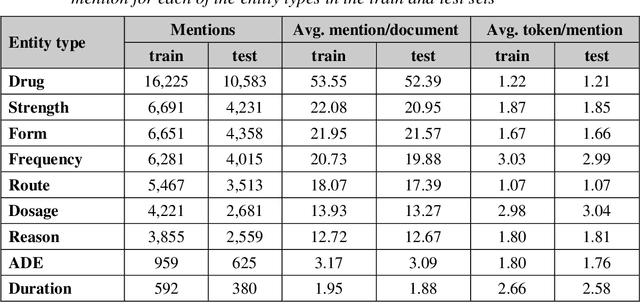
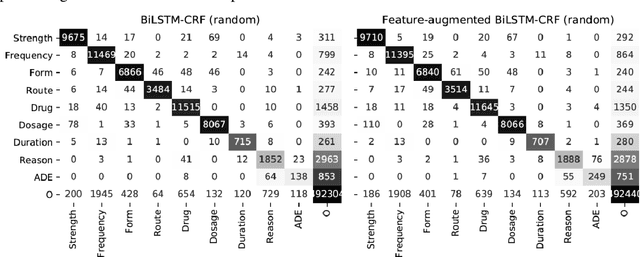
Abstract:Monitoring the administration of drugs and adverse drug reactions are key parts of pharmacovigilance. In this paper, we explore the extraction of drug mentions and drug-related information (reason for taking a drug, route, frequency, dosage, strength, form, duration, and adverse events) from hospital discharge summaries through deep learning that relies on various representations for clinical named entity recognition. This work was officially part of the 2018 n2c2 shared task, and we use the data supplied as part of the task. We developed two deep learning architecture based on recurrent neural networks and pre-trained language models. We also explore the effect of augmenting word representations with semantic features for clinical named entity recognition. Our feature-augmented BiLSTM-CRF model performed with F1-score of 92.67% and ranked 4th for entity extraction sub-task among submitted systems to n2c2 challenge. The recurrent neural networks that use the pre-trained domain-specific word embeddings and a CRF layer for label optimization perform drug, adverse event and related entities extraction with micro-averaged F1-score of over 91%. The augmentation of word vectors with semantic features extracted using available clinical NLP toolkits can further improve the performance. Word embeddings that are pre-trained on a large unannotated corpus of relevant documents and further fine-tuned to the task perform rather well. However, the augmentation of word embeddings with semantic features can help improve the performance (primarily by boosting precision) of drug-related named entity recognition from electronic health records.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge