H. R. Tizhoosh
Kimia Lab, University of Waterloo, Waterloo, ON, Canada, Vector Institute, MaRS Centre, Toronto, Canada
Colored Kimia Path24 Dataset: Configurations and Benchmarks with Deep Embeddings
Feb 15, 2021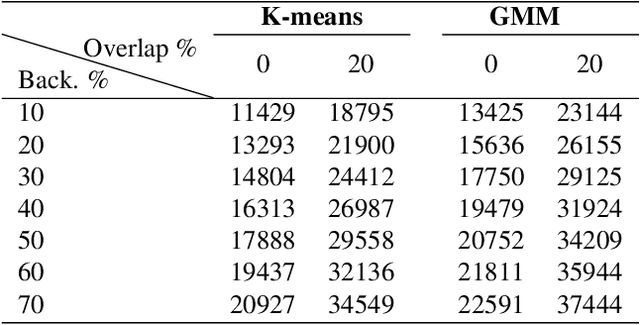
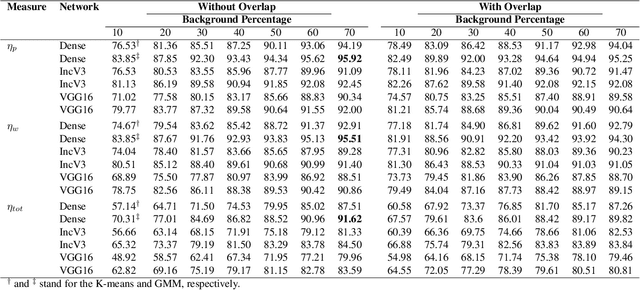

Abstract:The Kimia Path24 dataset has been introduced as a classification and retrieval dataset for digital pathology. Although it provides multi-class data, the color information has been neglected in the process of extracting patches. The staining information plays a major role in the recognition of tissue patterns. To address this drawback, we introduce the color version of Kimia Path24 by recreating sample patches from all 24 scans to propose Kimia Path24C. We run extensive experiments to determine the best configuration for selected patches. To provide preliminary results for setting a benchmark for the new dataset, we utilize VGG16, InceptionV3 and DenseNet-121 model as feature extractors. Then, we use these feature vectors to retrieve test patches. The accuracy of image retrieval using DenseNet was 95.92% while the highest accuracy using InceptionV3 and VGG16 reached 92.45% and 92%, respectively. We also experimented with "deep barcodes" and established that with a small loss in accuracy (e.g., 93.43% for binarized features for DenseNet instead of 95.92% when the features themselves are used), the search operations can be significantly accelerated.
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021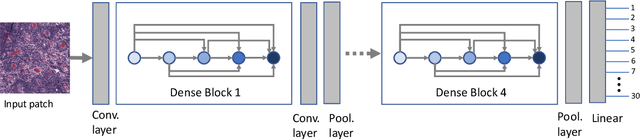
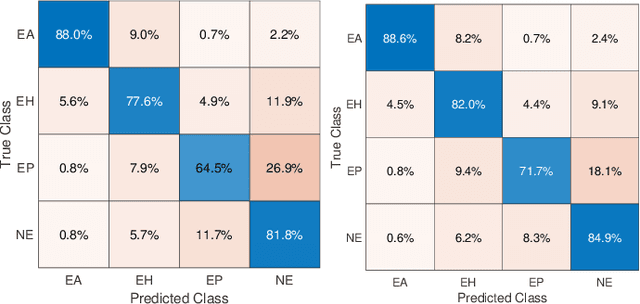
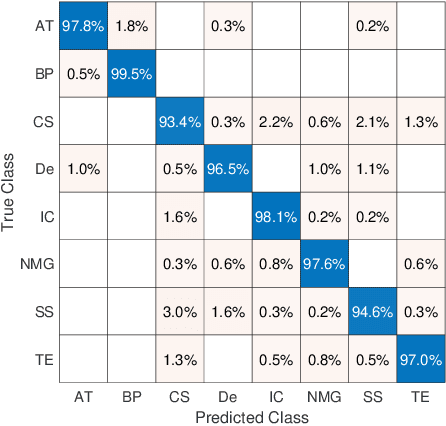
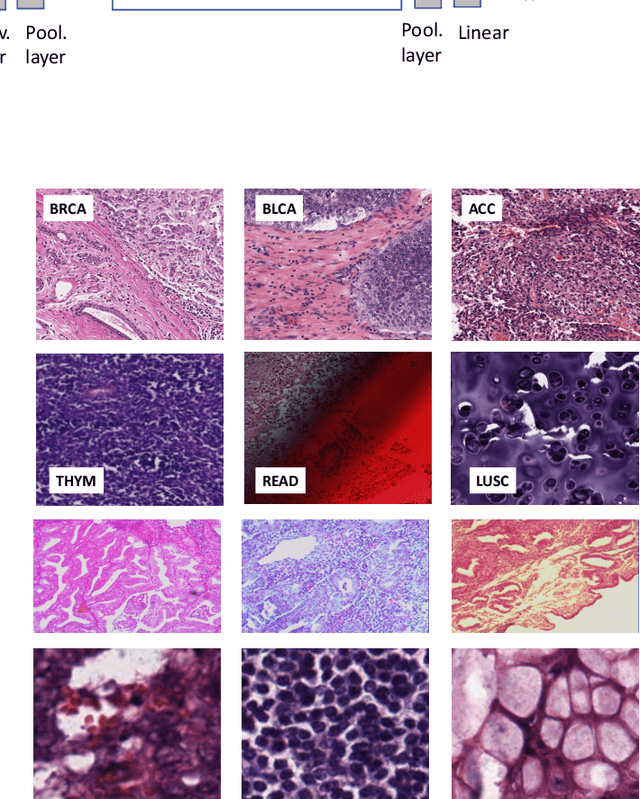
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
Magnification Generalization for Histopathology Image Embedding
Jan 18, 2021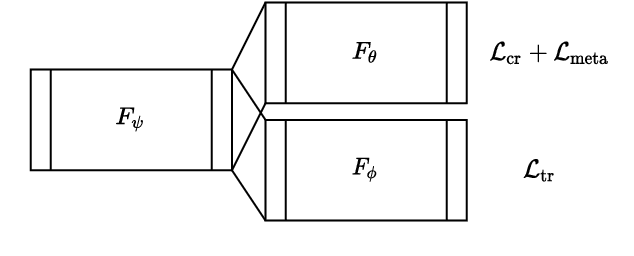
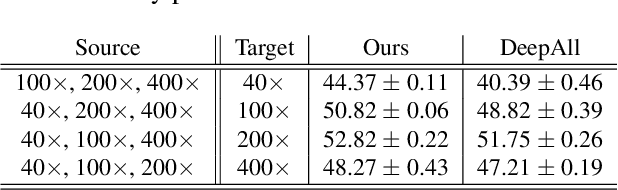
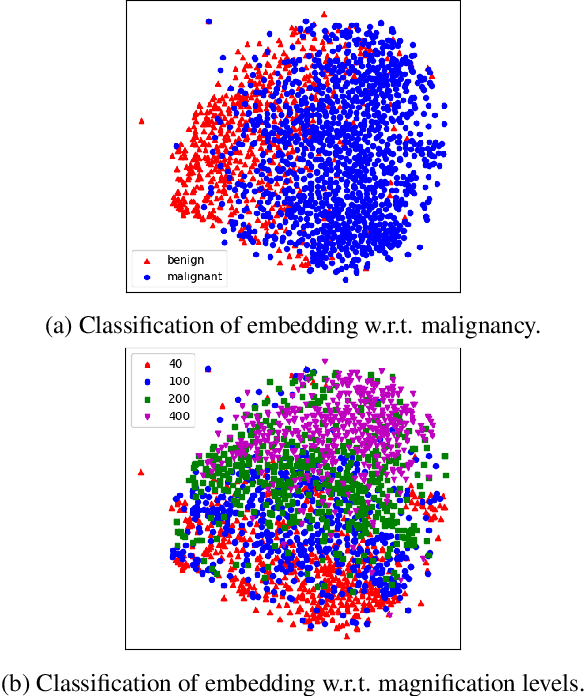
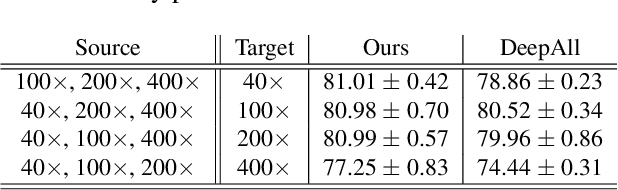
Abstract:Histopathology image embedding is an active research area in computer vision. Most of the embedding models exclusively concentrate on a specific magnification level. However, a useful task in histopathology embedding is to train an embedding space regardless of the magnification level. Two main approaches for tackling this goal are domain adaptation and domain generalization, where the target magnification levels may or may not be introduced to the model in training, respectively. Although magnification adaptation is a well-studied topic in the literature, this paper, to the best of our knowledge, is the first work on magnification generalization for histopathology image embedding. We use an episodic trainable domain generalization technique for magnification generalization, namely Model Agnostic Learning of Semantic Features (MASF), which works based on the Model Agnostic Meta-Learning (MAML) concept. Our experimental results on a breast cancer histopathology dataset with four different magnification levels show the proposed method's effectiveness for magnification generalization.
Ink Marker Segmentation in Histopathology Images Using Deep Learning
Oct 29, 2020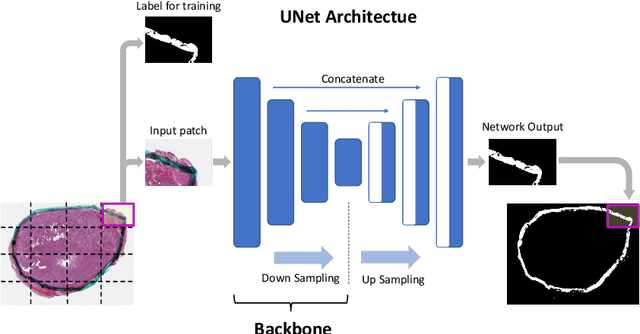
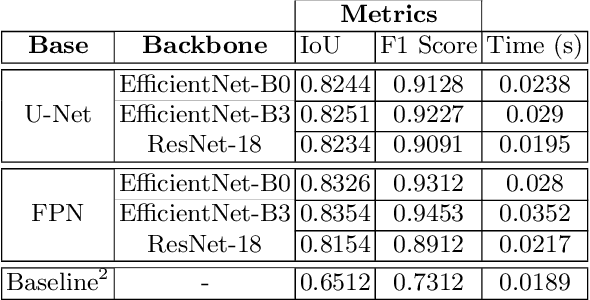
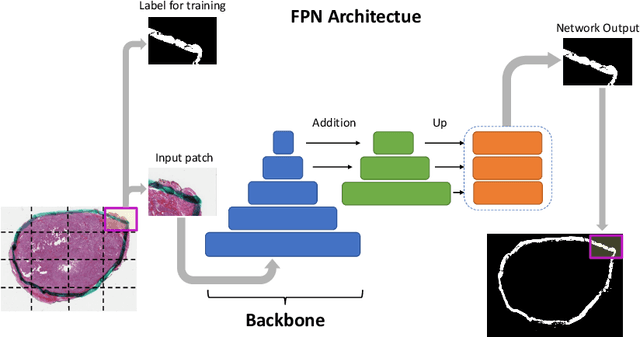
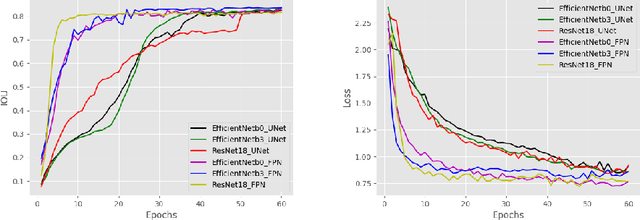
Abstract:Due to the recent advancements in machine vision, digital pathology has gained significant attention. Histopathology images are distinctly rich in visual information. The tissue glass slide images are utilized for disease diagnosis. Researchers study many methods to process histopathology images and facilitate fast and reliable diagnosis; therefore, the availability of high-quality slides becomes paramount. The quality of the images can be negatively affected when the glass slides are ink-marked by pathologists to delineate regions of interest. As an example, in one of the largest public histopathology datasets, The Cancer Genome Atlas (TCGA), approximately $12\%$ of the digitized slides are affected by manual delineations through ink markings. To process these open-access slide images and other repositories for the design and validation of new methods, an algorithm to detect the marked regions of the images is essential to avoid confusing tissue pixels with ink-colored pixels for computer methods. In this study, we propose to segment the ink-marked areas of pathology patches through a deep network. A dataset from $79$ whole slide images with $4,305$ patches was created and different networks were trained. Finally, the results showed an FPN model with the EffiecentNet-B3 as the backbone was found to be the superior configuration with an F1 score of $94.53\%$.
Batch-Incremental Triplet Sampling for Training Triplet Networks Using Bayesian Updating Theorem
Jul 10, 2020
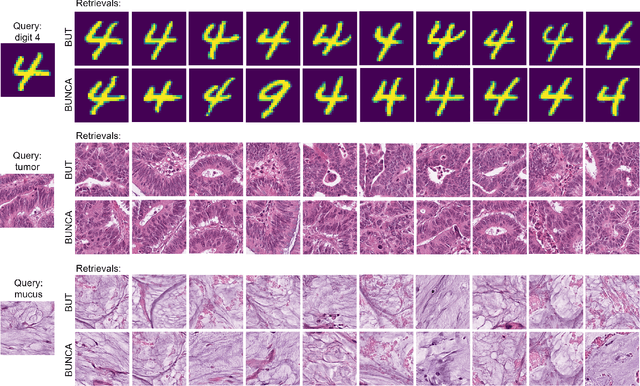
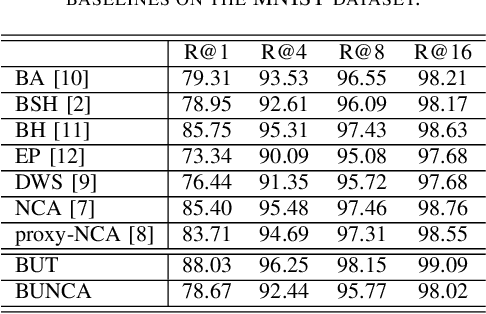
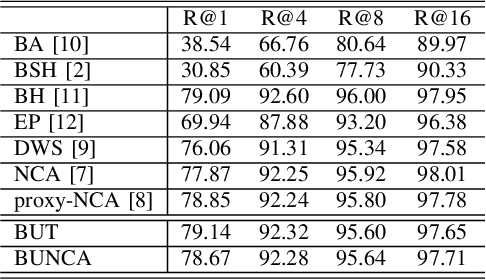
Abstract:Variants of Triplet networks are robust entities for learning a discriminative embedding subspace. There exist different triplet mining approaches for selecting the most suitable training triplets. Some of these mining methods rely on the extreme distances between instances, and some others make use of sampling. However, sampling from stochastic distributions of data rather than sampling merely from the existing embedding instances can provide more discriminative information. In this work, we sample triplets from distributions of data rather than from existing instances. We consider a multivariate normal distribution for the embedding of each class. Using Bayesian updating and conjugate priors, we update the distributions of classes dynamically by receiving the new mini-batches of training data. The proposed triplet mining with Bayesian updating can be used with any triplet-based loss function, e.g., triplet-loss or Neighborhood Component Analysis (NCA) loss. Accordingly, Our triplet mining approaches are called Bayesian Updating Triplet (BUT) and Bayesian Updating NCA (BUNCA), depending on which loss function is being used. Experimental results on two public datasets, namely MNIST and histopathology colorectal cancer (CRC), substantiate the effectiveness of the proposed triplet mining method.
Offline versus Online Triplet Mining based on Extreme Distances of Histopathology Patches
Jul 04, 2020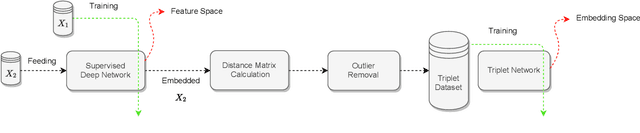

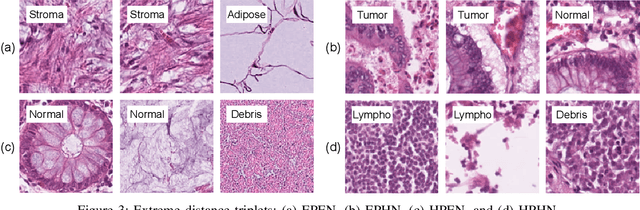
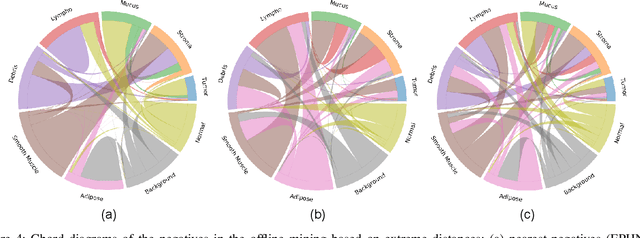
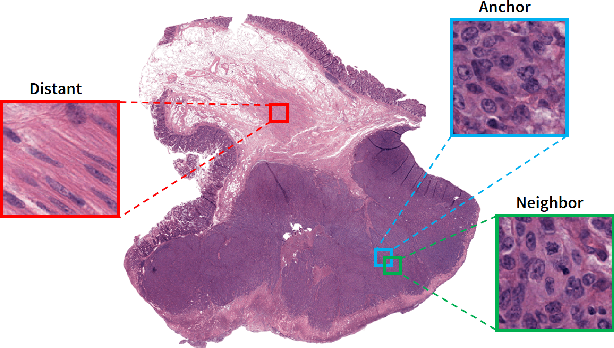
Abstract:We analyze the effect of offline and online triplet mining for colorectal cancer (CRC) histopathology dataset containing 100,000 patches. We consider the extreme, i.e., farthest and nearest patches with respect to a given anchor, both in online and offline mining. While many works focus solely on how to select the triplets online (batch-wise), we also study the effect of extreme distances and neighbor patches before training in an offline fashion. We analyze the impacts of extreme cases for offline versus online mining, including easy positive, batch semi-hard, and batch hard triplet mining as well as the neighborhood component analysis loss, its proxy version, and distance weighted sampling. We also investigate online approaches based on extreme distance and comprehensively compare the performance of offline and online mining based on the data patterns and explain offline mining as a tractable generalization of the online mining with large mini-batch size. As well, we discuss the relations of different colorectal tissue types in terms of extreme distances. We found that offline mining can generate a better statistical representation of the population by working on the whole dataset.
A Comparative Study of U-Net Topologies for Background Removal in Histopathology Images
Jun 08, 2020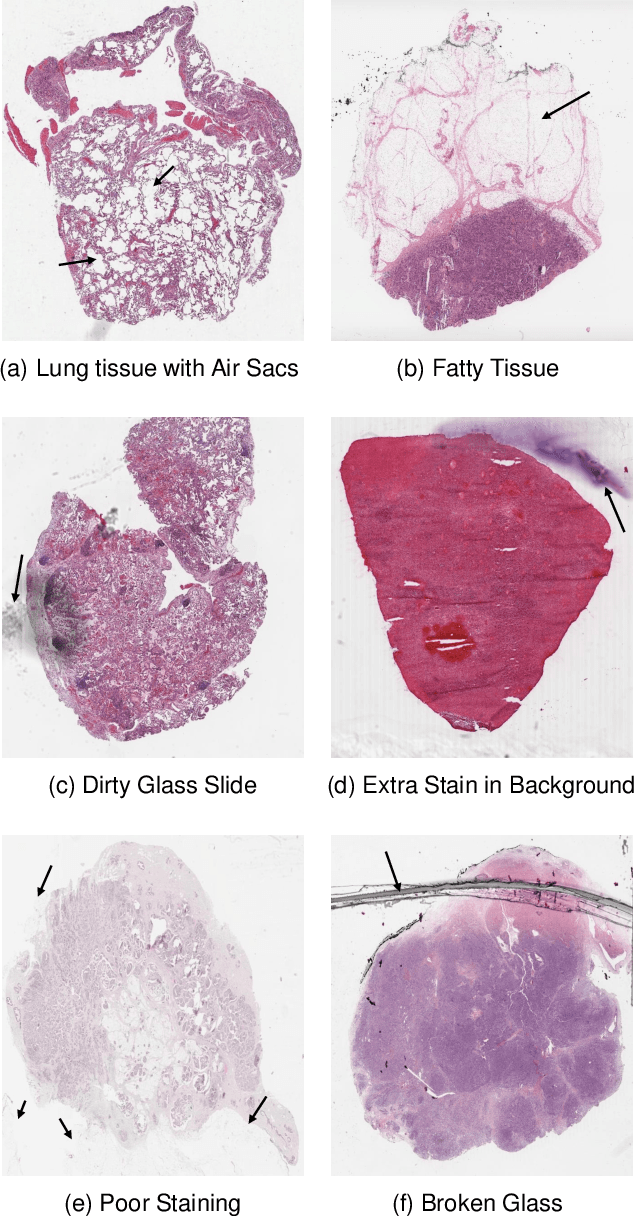
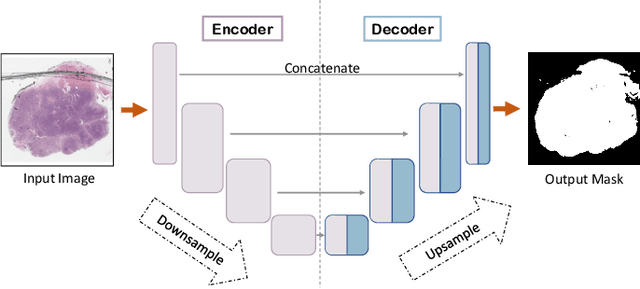
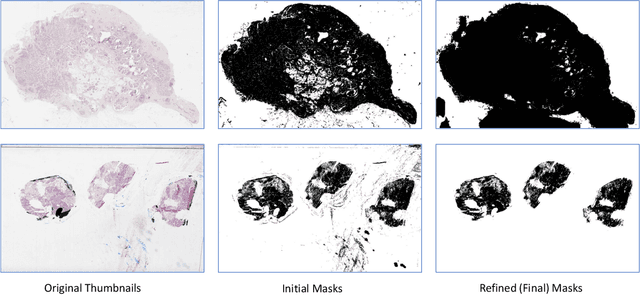
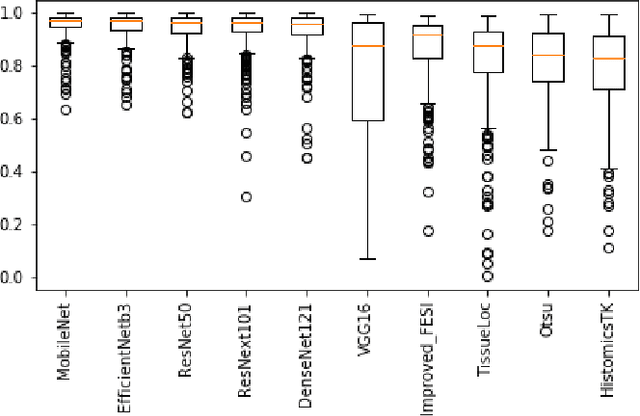
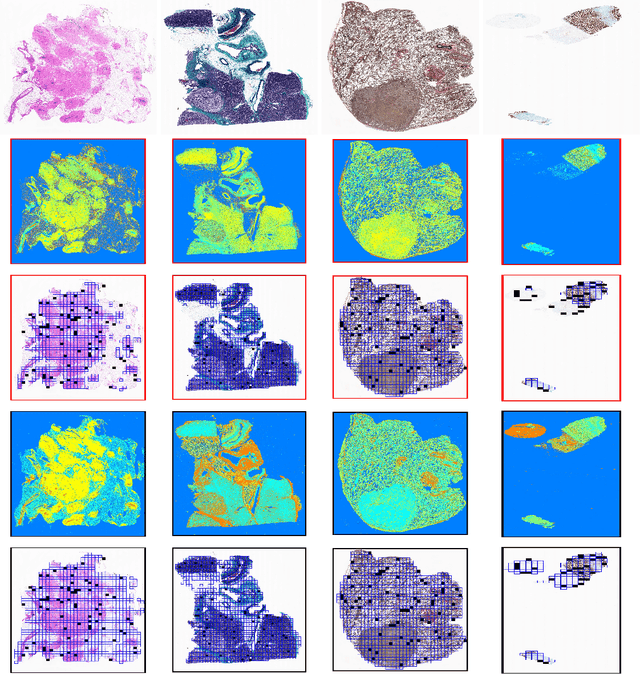
Abstract:During the last decade, the digitization of pathology has gained considerable momentum. Digital pathology offers many advantages including more efficient workflows, easier collaboration as well as a powerful venue for telepathology. At the same time, applying Computer-Aided Diagnosis (CAD) on Whole Slide Images (WSIs) has received substantial attention as a direct result of the digitization. The first step in any image analysis is to extract the tissue. Hence, background removal is an essential prerequisite for efficient and accurate results for many algorithms. In spite of the obvious discrimination for human operators, the identification of tissue regions in WSIs could be challenging for computers, mainly due to the existence of color variations and artifacts. Moreover, some cases such as alveolar tissue types, fatty tissues, and tissues with poor staining are difficult to detect. In this paper, we perform experiments on U-Net architecture with different network backbones (different topologies) to remove the background as well as artifacts from WSIs in order to extract the tissue regions. We compare a wide range of backbone networks including MobileNet, VGG16, EfficientNet-B3, ResNet50, ResNext101 and DenseNet121. We trained and evaluated the network on a manually labeled subset of The Cancer Genome Atlas (TCGA) Dataset. EfficientNet-B3 and MobileNet by almost 99% sensitivity and specificity reached the best results.
Supervision and Source Domain Impact on Representation Learning: A Histopathology Case Study
May 10, 2020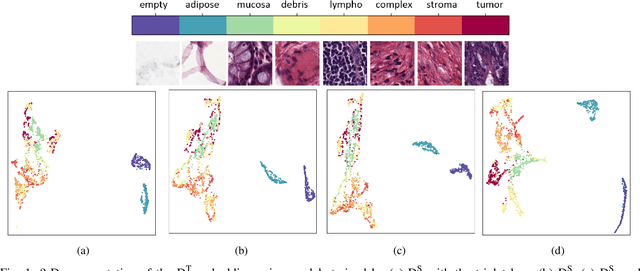

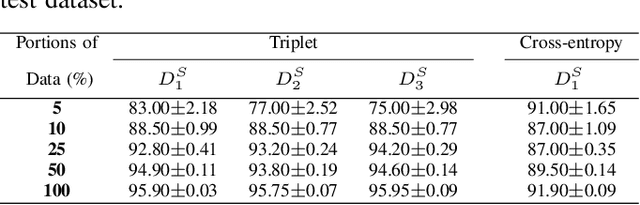
Abstract:As many algorithms depend on a suitable representation of data, learning unique features is considered a crucial task. Although supervised techniques using deep neural networks have boosted the performance of representation learning, the need for a large set of labeled data limits the application of such methods. As an example, high-quality delineations of regions of interest in the field of pathology is a tedious and time-consuming task due to the large image dimensions. In this work, we explored the performance of a deep neural network and triplet loss in the area of representation learning. We investigated the notion of similarity and dissimilarity in pathology whole-slide images and compared different setups from unsupervised and semi-supervised to supervised learning in our experiments. Additionally, different approaches were tested, applying few-shot learning on two publicly available pathology image datasets. We achieved high accuracy and generalization when the learned representations were applied to two different pathology datasets.
Recognizing Magnification Levels in Microscopic Snapshots
May 07, 2020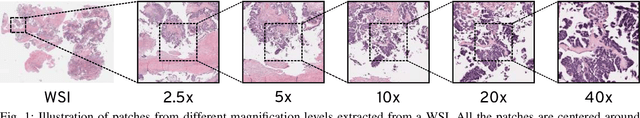
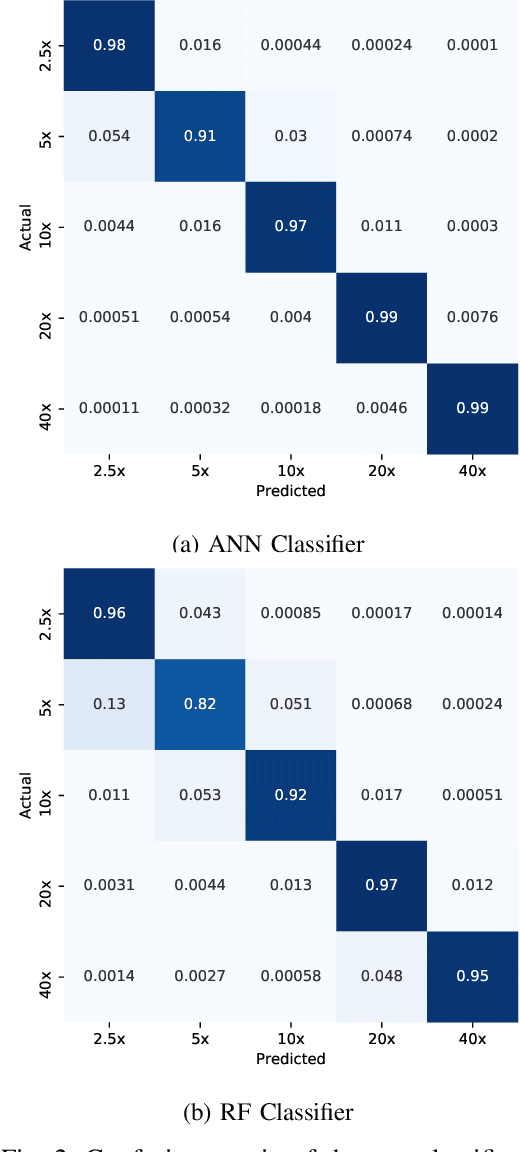
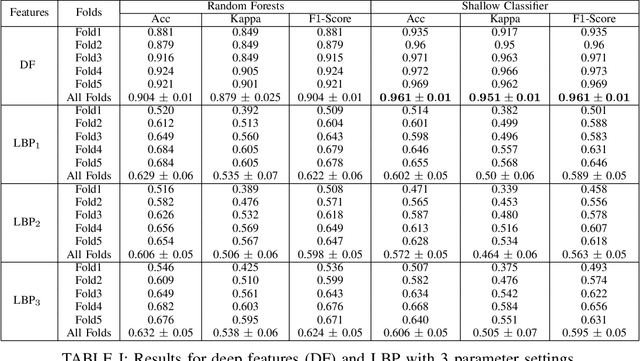
Abstract:Recent advances in digital imaging has transformed computer vision and machine learning to new tools for analyzing pathology images. This trend could automate some of the tasks in the diagnostic pathology and elevate the pathologist workload. The final step of any cancer diagnosis procedure is performed by the expert pathologist. These experts use microscopes with high level of optical magnification to observe minute characteristics of the tissue acquired through biopsy and fixed on glass slides. Switching between different magnifications, and finding the magnification level at which they identify the presence or absence of malignant tissues is important. As the majority of pathologists still use light microscopy, compared to digital scanners, in many instance a mounted camera on the microscope is used to capture snapshots from significant field-of-views. Repositories of such snapshots usually do not contain the magnification information. In this paper, we extract deep features of the images available on TCGA dataset with known magnification to train a classifier for magnification recognition. We compared the results with LBP, a well-known handcrafted feature extraction method. The proposed approach achieved a mean accuracy of 96% when a multi-layer perceptron was trained as a classifier.
Fisher Discriminant Triplet and Contrastive Losses for Training Siamese Networks
Apr 05, 2020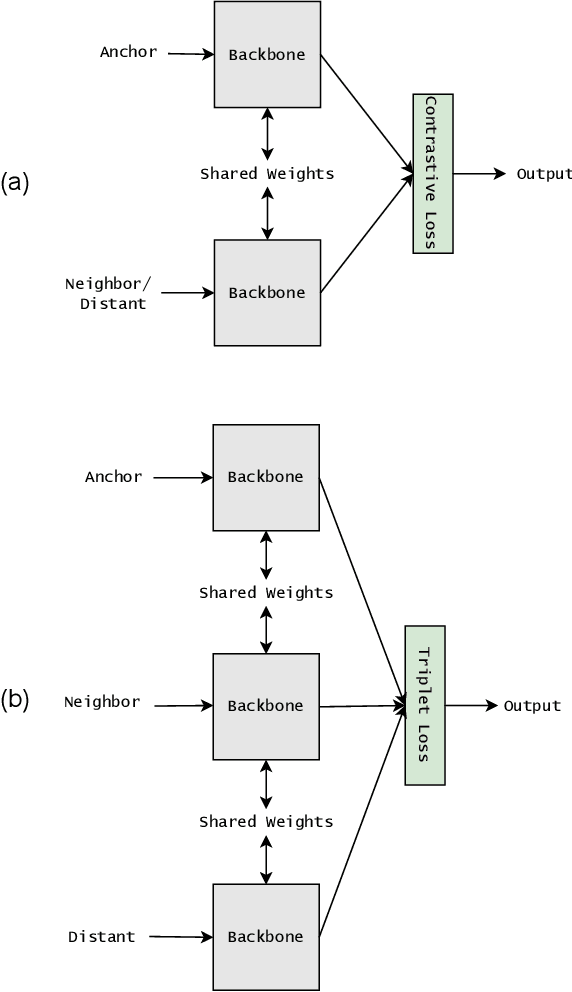
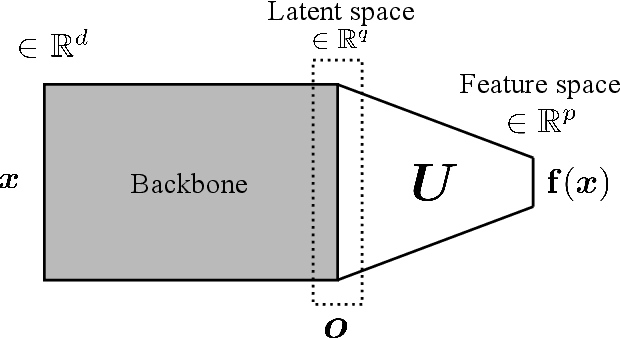
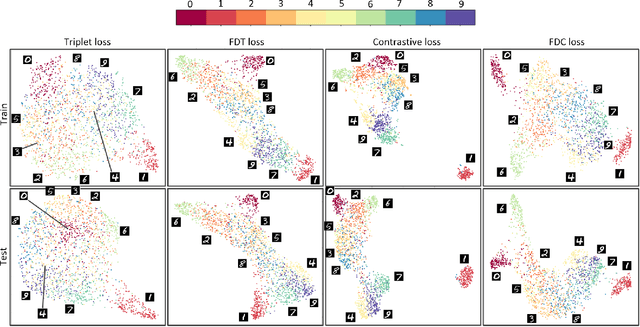
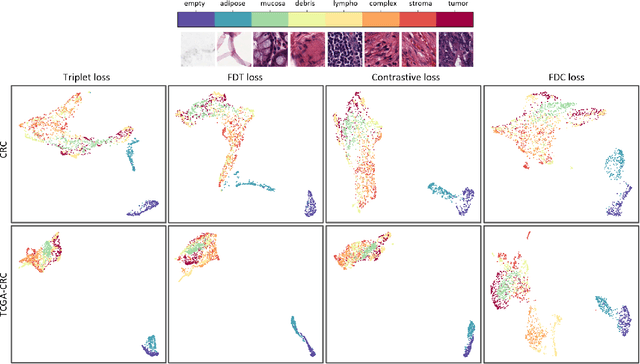
Abstract:Siamese neural network is a very powerful architecture for both feature extraction and metric learning. It usually consists of several networks that share weights. The Siamese concept is topology-agnostic and can use any neural network as its backbone. The two most popular loss functions for training these networks are the triplet and contrastive loss functions. In this paper, we propose two novel loss functions, named Fisher Discriminant Triplet (FDT) and Fisher Discriminant Contrastive (FDC). The former uses anchor-neighbor-distant triplets while the latter utilizes pairs of anchor-neighbor and anchor-distant samples. The FDT and FDC loss functions are designed based on the statistical formulation of the Fisher Discriminant Analysis (FDA), which is a linear subspace learning method. Our experiments on the MNIST and two challenging and publicly available histopathology datasets show the effectiveness of the proposed loss functions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge