Guoquan Pei
Omni-Fusion of Spatial and Spectral for Hyperspectral Image Segmentation
Jul 09, 2025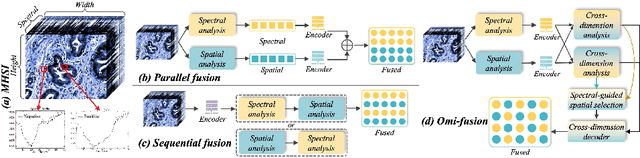
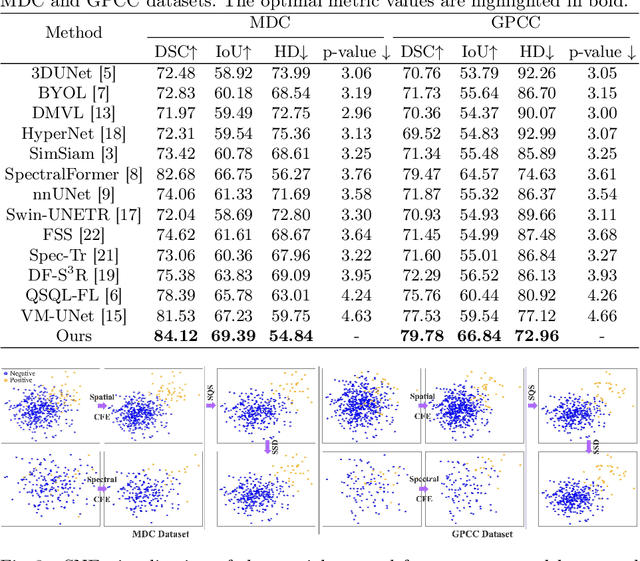
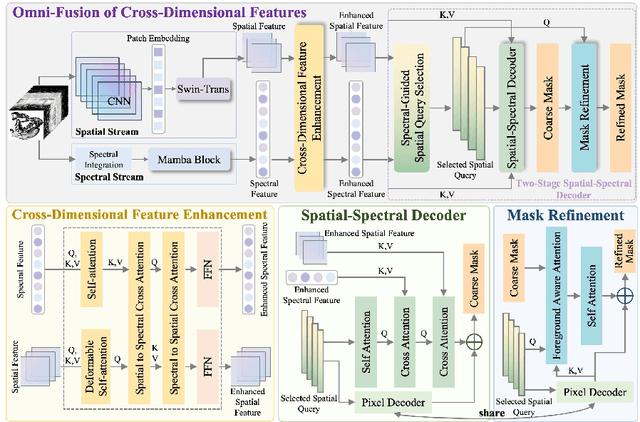
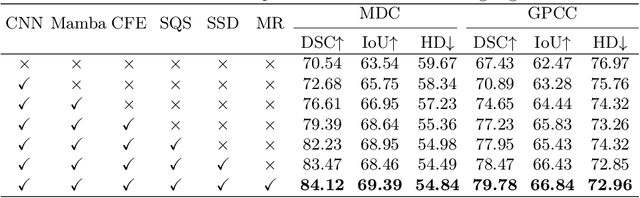
Abstract:Medical Hyperspectral Imaging (MHSI) has emerged as a promising tool for enhanced disease diagnosis, particularly in computational pathology, offering rich spectral information that aids in identifying subtle biochemical properties of tissues. Despite these advantages, effectively fusing both spatial-dimensional and spectral-dimensional information from MHSIs remains challenging due to its high dimensionality and spectral redundancy inherent characteristics. To solve the above challenges, we propose a novel spatial-spectral omni-fusion network for hyperspectral image segmentation, named as Omni-Fuse. Here, we introduce abundant cross-dimensional feature fusion operations, including a cross-dimensional enhancement module that refines both spatial and spectral features through bidirectional attention mechanisms, a spectral-guided spatial query selection to select the most spectral-related spatial feature as the query, and a two-stage cross-dimensional decoder which dynamically guide the model to focus on the selected spatial query. Despite of numerous attention blocks, Omni-Fuse remains efficient in execution. Experiments on two microscopic hyperspectral image datasets show that our approach can significantly improve the segmentation performance compared with the state-of-the-art methods, with over 5.73 percent improvement in DSC. Code available at: https://github.com/DeepMed-Lab-ECNU/Omni-Fuse.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge