Gabriele Lagani
Neuro-Inspired Visual Pattern Recognition via Biological Reservoir Computing
Feb 05, 2026Abstract:In this paper, we present a neuro-inspired approach to reservoir computing (RC) in which a network of in vitro cultured cortical neurons serves as the physical reservoir. Rather than relying on artificial recurrent models to approximate neural dynamics, our biological reservoir computing (BRC) system leverages the spontaneous and stimulus-evoked activity of living neural circuits as its computational substrate. A high-density multi-electrode array (HD-MEA) provides simultaneous stimulation and readout across hundreds of channels: input patterns are delivered through selected electrodes, while the remaining ones capture the resulting high-dimensional neural responses, yielding a biologically grounded feature representation. A linear readout layer (single-layer perceptron) is then trained to classify these reservoir states, enabling the living neural network to perform static visual pattern-recognition tasks within a computer-vision framework. We evaluate the system across a sequence of tasks of increasing difficulty, ranging from pointwise stimuli to oriented bars, clock-digit-like shapes, and handwritten digits from the MNIST dataset. Despite the inherent variability of biological neural responses-arising from noise, spontaneous activity, and inter-session differences-the system consistently generates high-dimensional representations that support accurate classification. These results demonstrate that in vitro cortical networks can function as effective reservoirs for static visual pattern recognition, opening new avenues for integrating living neural substrates into neuromorphic computing frameworks. More broadly, this work contributes to the effort to incorporate biological principles into machine learning and supports the goals of neuro-inspired vision by illustrating how living neural systems can inform the design of efficient and biologically grounded computational models.
CA3D: Convolutional-Attentional 3D Nets for Efficient Video Activity Recognition on the Edge
May 26, 2025Abstract:In this paper, we introduce a deep learning solution for video activity recognition that leverages an innovative combination of convolutional layers with a linear-complexity attention mechanism. Moreover, we introduce a novel quantization mechanism to further improve the efficiency of our model during both training and inference. Our model maintains a reduced computational cost, while preserving robust learning and generalization capabilities. Our approach addresses the issues related to the high computing requirements of current models, with the goal of achieving competitive accuracy on consumer and edge devices, enabling smart home and smart healthcare applications where efficiency and privacy issues are of concern. We experimentally validate our model on different established and publicly available video activity recognition benchmarks, improving accuracy over alternative models at a competitive computing cost.
From Neurons to Computation: Biological Reservoir Computing for Pattern Recognition
May 06, 2025Abstract:In this paper, we introduce a novel paradigm for reservoir computing (RC) that leverages a pool of cultured biological neurons as the reservoir substrate, creating a biological reservoir computing (BRC). This system operates similarly to an echo state network (ESN), with the key distinction that the neural activity is generated by a network of cultured neurons, rather than being modeled by traditional artificial computational units. The neuronal activity is recorded using a multi-electrode array (MEA), which enables high-throughput recording of neural signals. In our approach, inputs are introduced into the network through a subset of the MEA electrodes, while the remaining electrodes capture the resulting neural activity. This generates a nonlinear mapping of the input data to a high-dimensional biological feature space, where distinguishing between data becomes more efficient and straightforward, allowing a simple linear classifier to perform pattern recognition tasks effectively. To evaluate the performance of our proposed system, we present an experimental study that includes various input patterns, such as positional codes, bars with different orientations, and a digit recognition task. The results demonstrate the feasibility of using biological neural networks to perform tasks traditionally handled by artificial neural networks, paving the way for further exploration of biologically-inspired computing systems, with potential applications in neuromorphic engineering and bio-hybrid computing.
Comparison of Different Deep Neural Network Models in the Cultural Heritage Domain
Apr 30, 2025Abstract:The integration of computer vision and deep learning is an essential part of documenting and preserving cultural heritage, as well as improving visitor experiences. In recent years, two deep learning paradigms have been established in the field of computer vision: convolutional neural networks and transformer architectures. The present study aims to make a comparative analysis of some representatives of these two techniques of their ability to transfer knowledge from generic dataset, such as ImageNet, to cultural heritage specific tasks. The results of testing examples of the architectures VGG, ResNet, DenseNet, Visual Transformer, Swin Transformer, and PoolFormer, showed that DenseNet is the best in terms of efficiency-computability ratio.
Semi-Supervised Biomedical Image Segmentation via Diffusion Models and Teacher-Student Co-Training
Apr 02, 2025



Abstract:Supervised deep learning for semantic segmentation has achieved excellent results in accurately identifying anatomical and pathological structures in medical images. However, it often requires large annotated training datasets, which limits its scalability in clinical settings. To address this challenge, semi-supervised learning is a well-established approach that leverages both labeled and unlabeled data. In this paper, we introduce a novel semi-supervised teacher-student framework for biomedical image segmentation, inspired by the recent success of generative models. Our approach leverages denoising diffusion probabilistic models (DDPMs) to generate segmentation masks by progressively refining noisy inputs conditioned on the corresponding images. The teacher model is first trained in an unsupervised manner using a cycle-consistency constraint based on noise-corrupted image reconstruction, enabling it to generate informative semantic masks. Subsequently, the teacher is integrated into a co-training process with a twin-student network. The student learns from ground-truth labels when available and from teacher-generated pseudo-labels otherwise, while the teacher continuously improves its pseudo-labeling capabilities. Finally, to further enhance performance, we introduce a multi-round pseudo-label generation strategy that iteratively improves the pseudo-labeling process. We evaluate our approach on multiple biomedical imaging benchmarks, spanning multiple imaging modalities and segmentation tasks. Experimental results show that our method consistently outperforms state-of-the-art semi-supervised techniques, highlighting its effectiveness in scenarios with limited annotated data. The code to replicate our experiments can be found at https://github.com/ciampluca/diffusion_semi_supervised_biomedical_image_segmentation
Biologically-inspired Semi-supervised Semantic Segmentation for Biomedical Imaging
Dec 04, 2024



Abstract:We propose a novel two-stage semi-supervised learning approach for training downsampling-upsampling semantic segmentation architectures. The first stage does not use backpropagation. Rather, it exploits the bio-inspired Hebbian principle "fire together, wire together" as a local learning rule for updating the weights of both convolutional and transpose-convolutional layers, allowing unsupervised discovery of data features. In the second stage, the model is fine-tuned with standard backpropagation on a small subset of labeled data. We evaluate our methodology through experiments conducted on several widely used biomedical datasets, deeming that this domain is paramount in computer vision and is notably impacted by data scarcity. Results show that our proposed method outperforms SOTA approaches across different levels of label availability. Furthermore, we show that using our unsupervised stage to initialize the SOTA approaches leads to performance improvements. The code to replicate our experiments can be found at: https://github.com/ciampluca/hebbian-medical-image-segmentation
Spiking Neural Networks and Bio-Inspired Supervised Deep Learning: A Survey
Jul 30, 2023



Abstract:For a long time, biology and neuroscience fields have been a great source of inspiration for computer scientists, towards the development of Artificial Intelligence (AI) technologies. This survey aims at providing a comprehensive review of recent biologically-inspired approaches for AI. After introducing the main principles of computation and synaptic plasticity in biological neurons, we provide a thorough presentation of Spiking Neural Network (SNN) models, and we highlight the main challenges related to SNN training, where traditional backprop-based optimization is not directly applicable. Therefore, we discuss recent bio-inspired training methods, which pose themselves as alternatives to backprop, both for traditional and spiking networks. Bio-Inspired Deep Learning (BIDL) approaches towards advancing the computational capabilities and biological plausibility of current models.
Synaptic Plasticity Models and Bio-Inspired Unsupervised Deep Learning: A Survey
Jul 30, 2023



Abstract:Recently emerged technologies based on Deep Learning (DL) achieved outstanding results on a variety of tasks in the field of Artificial Intelligence (AI). However, these encounter several challenges related to robustness to adversarial inputs, ecological impact, and the necessity of huge amounts of training data. In response, researchers are focusing more and more interest on biologically grounded mechanisms, which are appealing due to the impressive capabilities exhibited by biological brains. This survey explores a range of these biologically inspired models of synaptic plasticity, their application in DL scenarios, and the connections with models of plasticity in Spiking Neural Networks (SNNs). Overall, Bio-Inspired Deep Learning (BIDL) represents an exciting research direction, aiming at advancing not only our current technologies but also our understanding of intelligence.
FastHebb: Scaling Hebbian Training of Deep Neural Networks to ImageNet Level
Jul 07, 2022
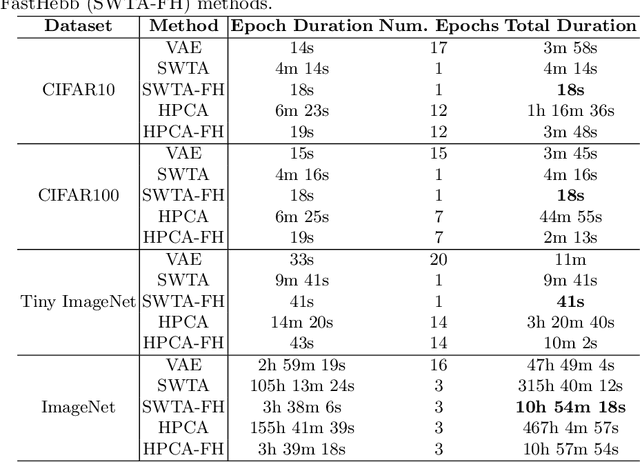

Abstract:Learning algorithms for Deep Neural Networks are typically based on supervised end-to-end Stochastic Gradient Descent (SGD) training with error backpropagation (backprop). Backprop algorithms require a large number of labelled training samples to achieve high performance. However, in many realistic applications, even if there is plenty of image samples, very few of them are labelled, and semi-supervised sample-efficient training strategies have to be used. Hebbian learning represents a possible approach towards sample efficient training; however, in current solutions, it does not scale well to large datasets. In this paper, we present FastHebb, an efficient and scalable solution for Hebbian learning which achieves higher efficiency by 1) merging together update computation and aggregation over a batch of inputs, and 2) leveraging efficient matrix multiplication algorithms on GPU. We validate our approach on different computer vision benchmarks, in a semi-supervised learning scenario. FastHebb outperforms previous solutions by up to 50 times in terms of training speed, and notably, for the first time, we are able to bring Hebbian algorithms to ImageNet scale.
Deep Features for CBIR with Scarce Data using Hebbian Learning
May 18, 2022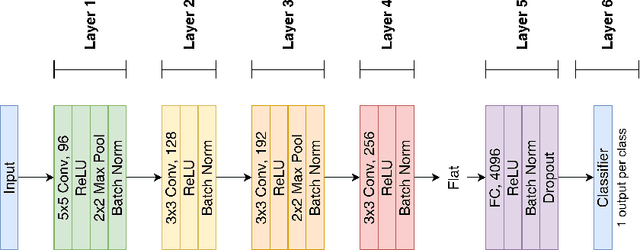
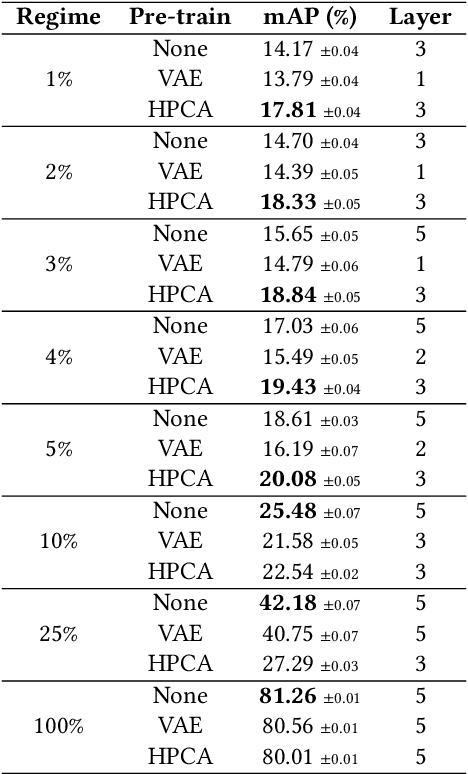
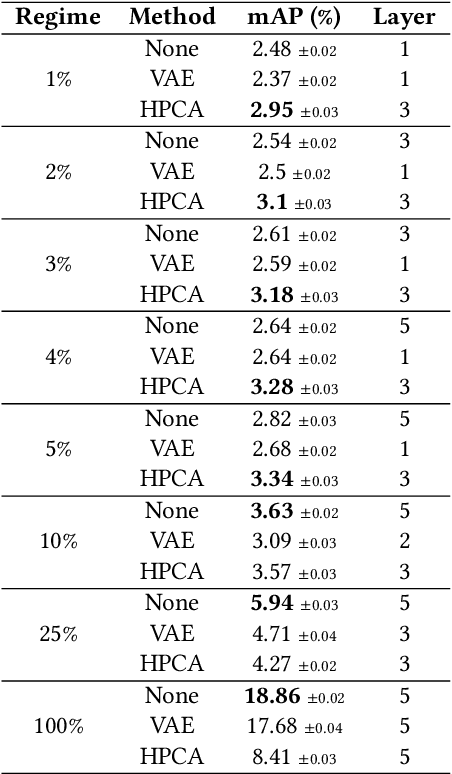
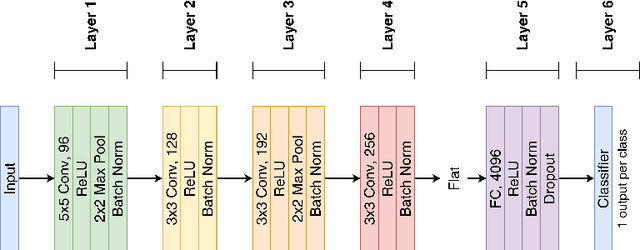
Abstract:Features extracted from Deep Neural Networks (DNNs) have proven to be very effective in the context of Content Based Image Retrieval (CBIR). In recent work, biologically inspired \textit{Hebbian} learning algorithms have shown promises for DNN training. In this contribution, we study the performance of such algorithms in the development of feature extractors for CBIR tasks. Specifically, we consider a semi-supervised learning strategy in two steps: first, an unsupervised pre-training stage is performed using Hebbian learning on the image dataset; second, the network is fine-tuned using supervised Stochastic Gradient Descent (SGD) training. For the unsupervised pre-training stage, we explore the nonlinear Hebbian Principal Component Analysis (HPCA) learning rule. For the supervised fine-tuning stage, we assume sample efficiency scenarios, in which the amount of labeled samples is just a small fraction of the whole dataset. Our experimental analysis, conducted on the CIFAR10 and CIFAR100 datasets shows that, when few labeled samples are available, our Hebbian approach provides relevant improvements compared to various alternative methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge