Francois-Xavier Vialard
multiGradICON: A Foundation Model for Multimodal Medical Image Registration
Aug 01, 2024



Abstract:Modern medical image registration approaches predict deformations using deep networks. These approaches achieve state-of-the-art (SOTA) registration accuracy and are generally fast. However, deep learning (DL) approaches are, in contrast to conventional non-deep-learning-based approaches, anatomy-specific. Recently, a universal deep registration approach, uniGradICON, has been proposed. However, uniGradICON focuses on monomodal image registration. In this work, we therefore develop multiGradICON as a first step towards universal *multimodal* medical image registration. Specifically, we show that 1) we can train a DL registration model that is suitable for monomodal *and* multimodal registration; 2) loss function randomization can increase multimodal registration accuracy; and 3) training a model with multimodal data helps multimodal generalization. Our code and the multiGradICON model are available at https://github.com/uncbiag/uniGradICON.
CARL: A Framework for Equivariant Image Registration
May 28, 2024



Abstract:Image registration estimates spatial correspondences between a pair of images. These estimates are typically obtained via numerical optimization or regression by a deep network. A desirable property of such estimators is that a correspondence estimate (e.g., the true oracle correspondence) for an image pair is maintained under deformations of the input images. Formally, the estimator should be equivariant to a desired class of image transformations. In this work, we present careful analyses of the desired equivariance properties in the context of multi-step deep registration networks. Based on these analyses we 1) introduce the notions of $[U,U]$ equivariance (network equivariance to the same deformations of the input images) and $[W,U]$ equivariance (where input images can undergo different deformations); we 2) show that in a suitable multi-step registration setup it is sufficient for overall $[W,U]$ equivariance if the first step has $[W,U]$ equivariance and all others have $[U,U]$ equivariance; we 3) show that common displacement-predicting networks only exhibit $[U,U]$ equivariance to translations instead of the more powerful $[W,U]$ equivariance; and we 4) show how to achieve multi-step $[W,U]$ equivariance via a coordinate-attention mechanism combined with displacement-predicting refinement layers (CARL). Overall, our approach obtains excellent practical registration performance on several 3D medical image registration tasks and outperforms existing unsupervised approaches for the challenging problem of abdomen registration.
uniGradICON: A Foundation Model for Medical Image Registration
Mar 09, 2024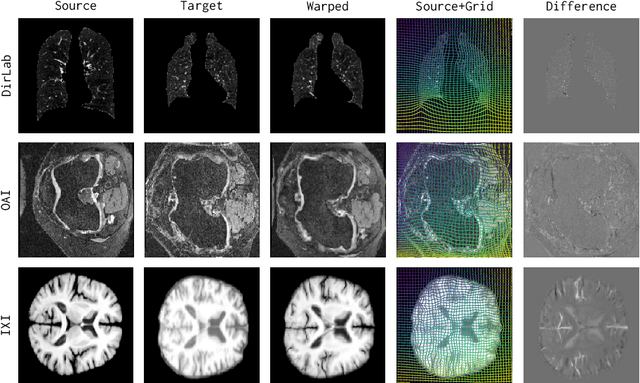
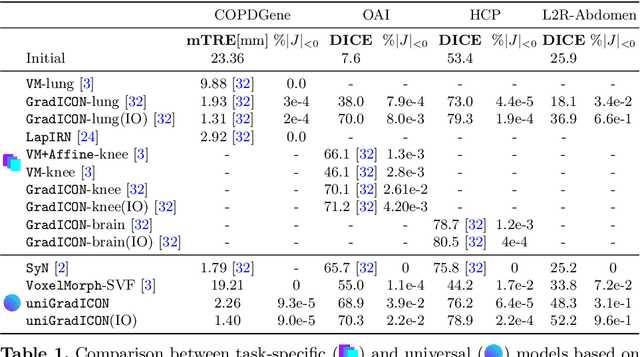

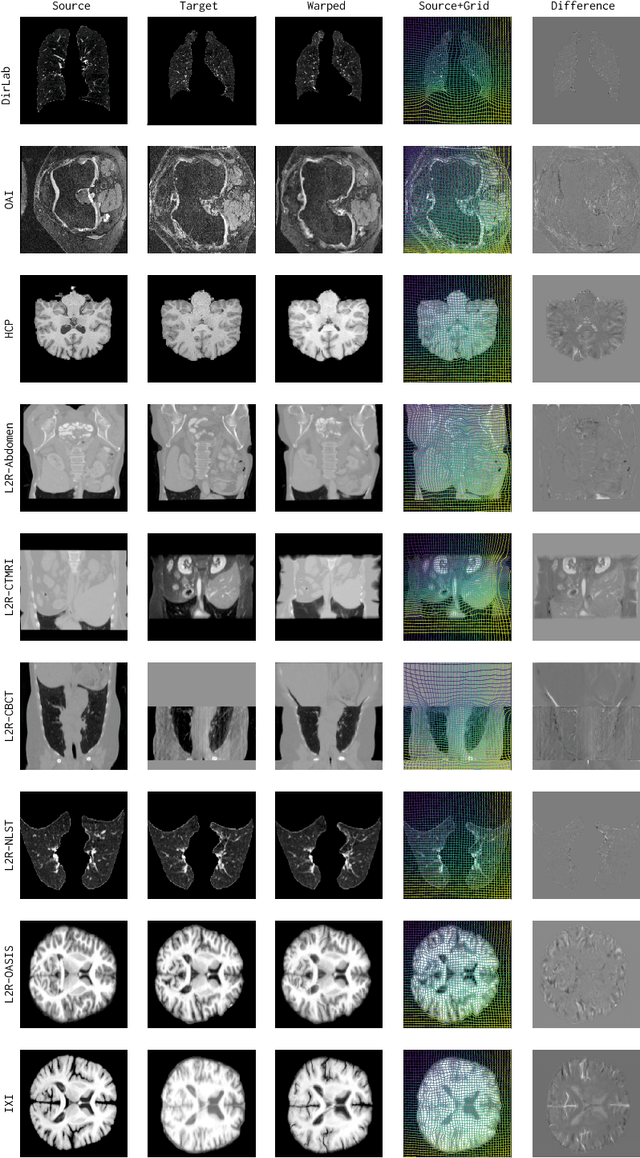
Abstract:Conventional medical image registration approaches directly optimize over the parameters of a transformation model. These approaches have been highly successful and are used generically for registrations of different anatomical regions. Recent deep registration networks are incredibly fast and accurate but are only trained for specific tasks. Hence, they are no longer generic registration approaches. We therefore propose uniGradICON, a first step toward a foundation model for registration providing 1) great performance \emph{across} multiple datasets which is not feasible for current learning-based registration methods, 2) zero-shot capabilities for new registration tasks suitable for different acquisitions, anatomical regions, and modalities compared to the training dataset, and 3) a strong initialization for finetuning on out-of-distribution registration tasks. UniGradICON unifies the speed and accuracy benefits of learning-based registration algorithms with the generic applicability of conventional non-deep-learning approaches. We extensively trained and evaluated uniGradICON on twelve different public datasets. Our code and the uniGradICON model are available at https://github.com/uncbiag/uniGradICON.
Inverse Consistency by Construction for Multistep Deep Registration
Apr 28, 2023Abstract:Inverse consistency is a desirable property for image registration. We propose a simple technique to make a neural registration network inverse consistent by construction, as a consequence of its structure, as long as it parameterizes its output transform by a Lie group. We extend this technique to multi-step neural registration by composing many such networks in a way that preserves inverse consistency. This multi-step approach also allows for inverse-consistent coarse to fine registration. We evaluate our technique on synthetic 2-D data and four 3-D medical image registration tasks and obtain excellent registration accuracy while assuring inverse consistency.
ICON: Learning Regular Maps Through Inverse Consistency
May 21, 2021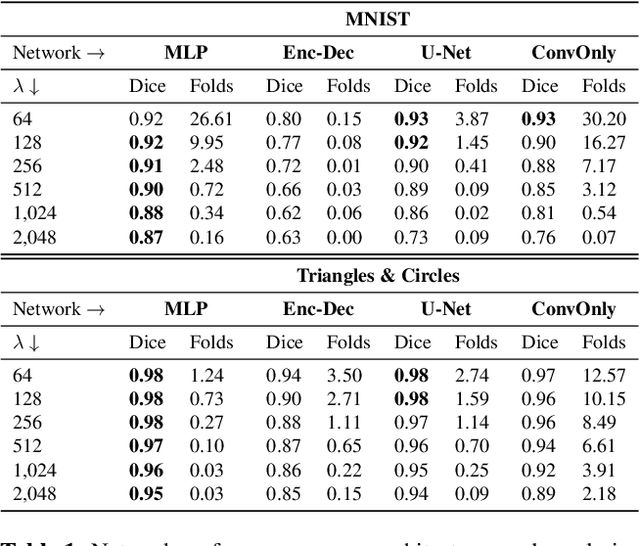
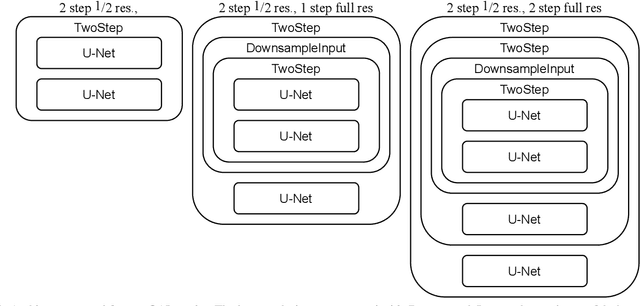
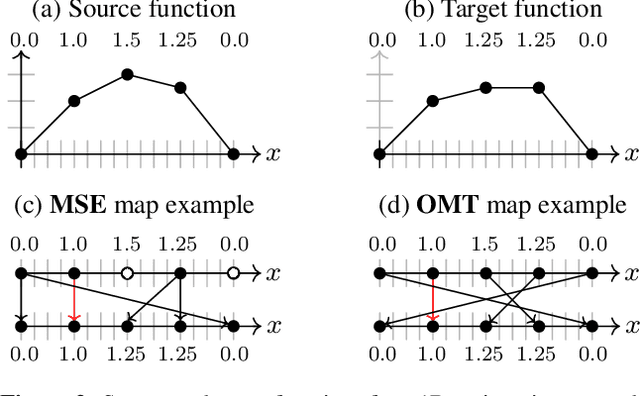
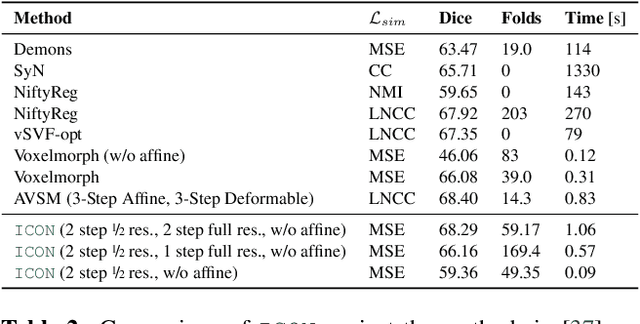
Abstract:Learning maps between data samples is fundamental. Applications range from representation learning, image translation and generative modeling, to the estimation of spatial deformations. Such maps relate feature vectors, or map between feature spaces. Well-behaved maps should be regular, which can be imposed explicitly or may emanate from the data itself. We explore what induces regularity for spatial transformations, e.g., when computing image registrations. Classical optimization-based models compute maps between pairs of samples and rely on an appropriate regularizer for well-posedness. Recent deep learning approaches have attempted to avoid using such regularizers altogether by relying on the sample population instead. We explore if it is possible to obtain spatial regularity using an inverse consistency loss only and elucidate what explains map regularity in such a context. We find that deep networks combined with an inverse consistency loss and randomized off-grid interpolation yield well behaved, approximately diffeomorphic, spatial transformations. Despite the simplicity of this approach, our experiments present compelling evidence, on both synthetic and real data, that regular maps can be obtained without carefully tuned explicit regularizers, while achieving competitive registration performance.
Metric Learning for Image Registration
Apr 21, 2019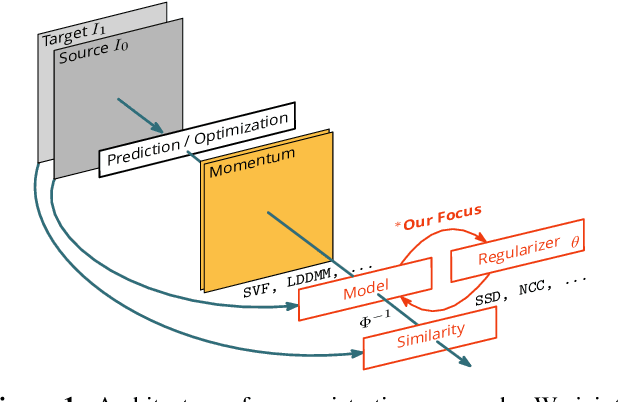
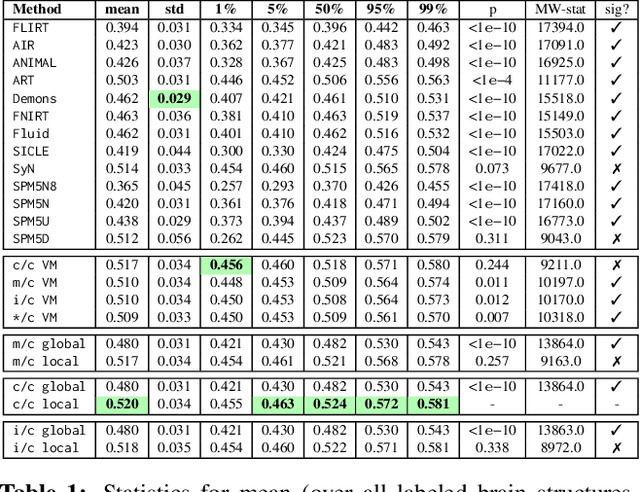
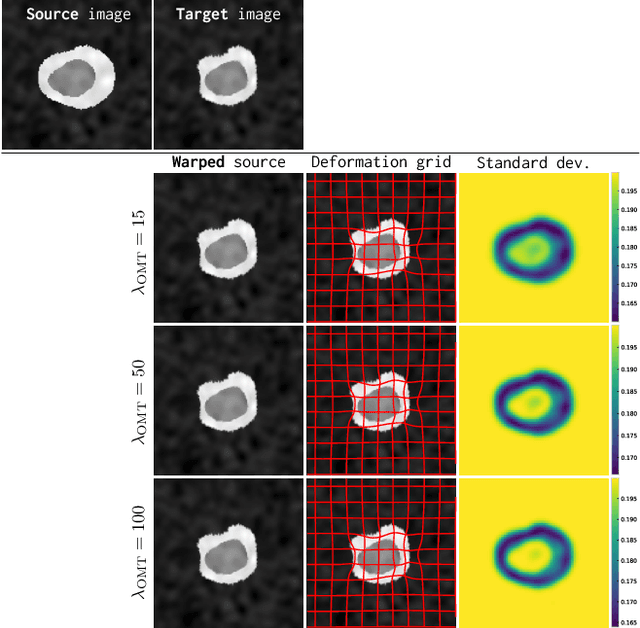
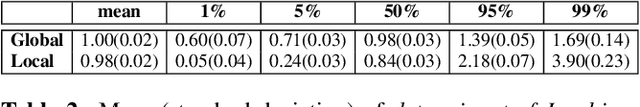
Abstract:Image registration is a key technique in medical image analysis to estimate deformations between image pairs. A good deformation model is important for high-quality estimates. However, most existing approaches use ad-hoc deformation models chosen for mathematical convenience rather than to capture observed data variation. Recent deep learning approaches learn deformation models directly from data. However, they provide limited control over the spatial regularity of transformations. Instead of learning the entire registration approach, we learn a spatially-adaptive regularizer within a registration model. This allows controlling the desired level of regularity and preserving structural properties of a registration model. For example, diffeomorphic transformations can be attained. Our approach is a radical departure from existing deep learning approaches to image registration by embedding a deep learning model in an optimization-based registration algorithm to parameterize and data-adapt the registration model itself.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge