Felix Wong
MONET -- Virtual Cell Painting of Brightfield Images and Time Lapses Using Reference Consistent Diffusion
Dec 12, 2025Abstract:Cell painting is a popular technique for creating human-interpretable, high-contrast images of cell morphology. There are two major issues with cell paint: (1) it is labor-intensive and (2) it requires chemical fixation, making the study of cell dynamics impossible. We train a diffusion model (Morphological Observation Neural Enhancement Tool, or MONET) on a large dataset to predict cell paint channels from brightfield images. We show that model quality improves with scale. The model uses a consistency architecture to generate time-lapse videos, despite the impossibility of obtaining cell paint video training data. In addition, we show that this architecture enables a form of in-context learning, allowing the model to partially transfer to out-of-distribution cell lines and imaging protocols. Virtual cell painting is not intended to replace physical cell painting completely, but to act as a complementary tool enabling novel workflows in biological research.
From which world is your graph?
Nov 03, 2017
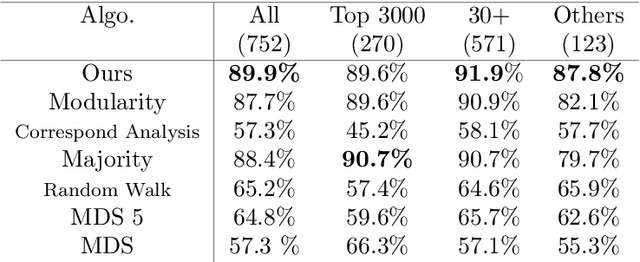
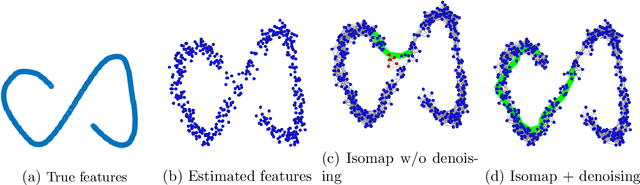
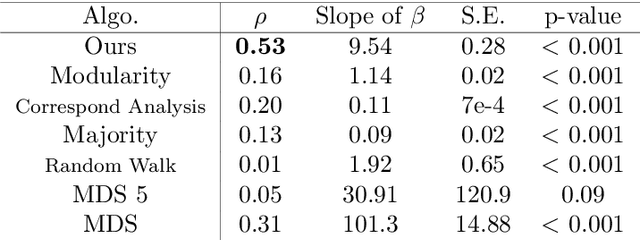
Abstract:Discovering statistical structure from links is a fundamental problem in the analysis of social networks. Choosing a misspecified model, or equivalently, an incorrect inference algorithm will result in an invalid analysis or even falsely uncover patterns that are in fact artifacts of the model. This work focuses on unifying two of the most widely used link-formation models: the stochastic blockmodel (SBM) and the small world (or latent space) model (SWM). Integrating techniques from kernel learning, spectral graph theory, and nonlinear dimensionality reduction, we develop the first statistically sound polynomial-time algorithm to discover latent patterns in sparse graphs for both models. When the network comes from an SBM, the algorithm outputs a block structure. When it is from an SWM, the algorithm outputs estimates of each node's latent position.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge