Erin Craig
Supervised learning pays attention
Dec 10, 2025Abstract:In-context learning with attention enables large neural networks to make context-specific predictions by selectively focusing on relevant examples. Here, we adapt this idea to supervised learning procedures such as lasso regression and gradient boosting, for tabular data. Our goals are to (1) flexibly fit personalized models for each prediction point and (2) retain model simplicity and interpretability. Our method fits a local model for each test observation by weighting the training data according to attention, a supervised similarity measure that emphasizes features and interactions that are predictive of the outcome. Attention weighting allows the method to adapt to heterogeneous data in a data-driven way, without requiring cluster or similarity pre-specification. Further, our approach is uniquely interpretable: for each test observation, we identify which features are most predictive and which training observations are most relevant. We then show how to use attention weighting for time series and spatial data, and we present a method for adapting pretrained tree-based models to distributional shift using attention-weighted residual corrections. Across real and simulated datasets, attention weighting improves predictive performance while preserving interpretability, and theory shows that attention-weighting linear models attain lower mean squared error than the standard linear model under mixture-of-models data-generating processes with known subgroup structure.
MMIL: A novel algorithm for disease associated cell type discovery
Jun 12, 2024



Abstract:Single-cell datasets often lack individual cell labels, making it challenging to identify cells associated with disease. To address this, we introduce Mixture Modeling for Multiple Instance Learning (MMIL), an expectation maximization method that enables the training and calibration of cell-level classifiers using patient-level labels. Our approach can be used to train e.g. lasso logistic regression models, gradient boosted trees, and neural networks. When applied to clinically-annotated, primary patient samples in Acute Myeloid Leukemia (AML) and Acute Lymphoblastic Leukemia (ALL), our method accurately identifies cancer cells, generalizes across tissues and treatment timepoints, and selects biologically relevant features. In addition, MMIL is capable of incorporating cell labels into model training when they are known, providing a powerful framework for leveraging both labeled and unlabeled data simultaneously. Mixture Modeling for MIL offers a novel approach for cell classification, with significant potential to advance disease understanding and management, especially in scenarios with unknown gold-standard labels and high dimensionality.
Using Pre-training and Interaction Modeling for ancestry-specific disease prediction in UK Biobank
Apr 26, 2024



Abstract:Recent genome-wide association studies (GWAS) have uncovered the genetic basis of complex traits, but show an under-representation of non-European descent individuals, underscoring a critical gap in genetic research. Here, we assess whether we can improve disease prediction across diverse ancestries using multiomic data. We evaluate the performance of Group-LASSO INTERaction-NET (glinternet) and pretrained lasso in disease prediction focusing on diverse ancestries in the UK Biobank. Models were trained on data from White British and other ancestries and validated across a cohort of over 96,000 individuals for 8 diseases. Out of 96 models trained, we report 16 with statistically significant incremental predictive performance in terms of ROC-AUC scores. These findings suggest that advanced statistical methods that borrow information across multiple ancestries may improve disease risk prediction, but with limited benefit.
Predicting readmission risk from doctors' notes
Dec 20, 2017
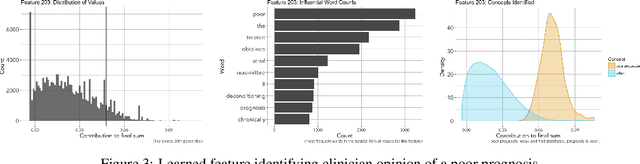
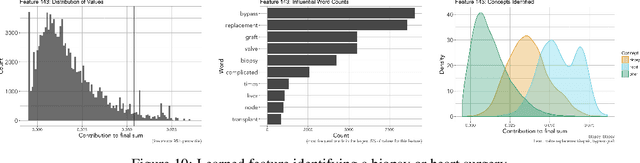
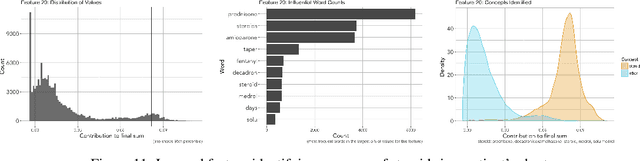
Abstract:We develop a model using deep learning techniques and natural language processing on unstructured text from medical records to predict hospital-wide $30$-day unplanned readmission, with c-statistic $.70$. Our model is constructed to allow physicians to interpret the significant features for prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge