Eldad Klaiman
B-Cos Aligned Transformers Learn Human-Interpretable Features
Jan 18, 2024Abstract:Vision Transformers (ViTs) and Swin Transformers (Swin) are currently state-of-the-art in computational pathology. However, domain experts are still reluctant to use these models due to their lack of interpretability. This is not surprising, as critical decisions need to be transparent and understandable. The most common approach to understanding transformers is to visualize their attention. However, attention maps of ViTs are often fragmented, leading to unsatisfactory explanations. Here, we introduce a novel architecture called the B-cos Vision Transformer (BvT) that is designed to be more interpretable. It replaces all linear transformations with the B-cos transform to promote weight-input alignment. In a blinded study, medical experts clearly ranked BvTs above ViTs, suggesting that our network is better at capturing biomedically relevant structures. This is also true for the B-cos Swin Transformer (Bwin). Compared to the Swin Transformer, it even improves the F1-score by up to 4.7% on two public datasets.
Training Transitive and Commutative Multimodal Transformers with LoReTTa
May 23, 2023Abstract:Collecting a multimodal dataset with two paired modalities A and B or B and C is difficult in practice. Obtaining a dataset with three aligned modalities A, B, and C is even more challenging. For example, some public medical datasets have only genetic sequences and microscopic images for one patient, and only genetic sequences and radiological images for another - but no dataset includes both microscopic and radiological images for the same patient. This makes it difficult to integrate and combine all modalities into a large pre-trained neural network. We introduce LoReTTa (Linking mOdalities with a tRansitive and commutativE pre-Training sTrAtegy) to address this understudied problem. Our self-supervised framework combines causal masked modeling with the rules of commutativity and transitivity to transition within and between different modalities. Thus, it can model the relation A -> C with A -> B -> C. Given a dataset containing only the disjoint combinations (A, B) and (B, C), we show that a transformer pre-trained with LoReTTa can handle any modality combination at inference time, including the never-seen pair (A, C) and the triplet (A, B, C). We evaluate our approach on a multimodal dataset derived from MNIST containing speech, vision, and language, as well as a real-world medical dataset containing mRNA, miRNA, and RPPA samples from TCGA. Compared to traditional pre-training methods, we observe up to a 100-point reduction in perplexity for autoregressive generation tasks and up to a 15% improvement in classification accuracy for previously unseen modality pairs during the pre-training phase.
HEROHE Challenge: assessing HER2 status in breast cancer without immunohistochemistry or in situ hybridization
Nov 08, 2021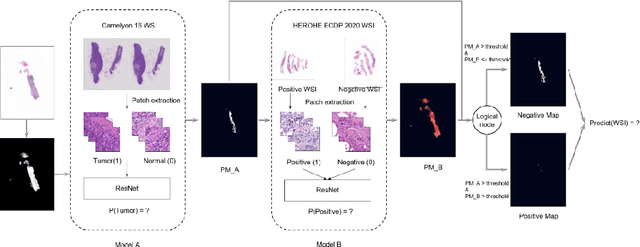
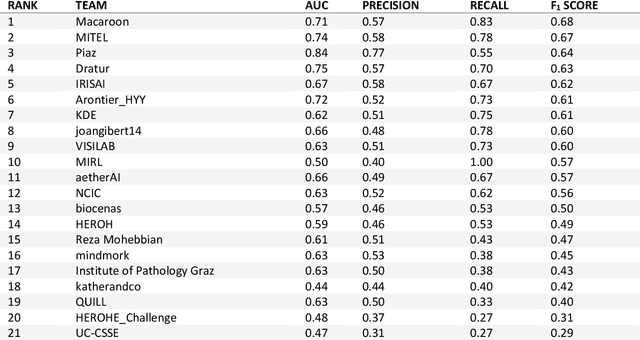
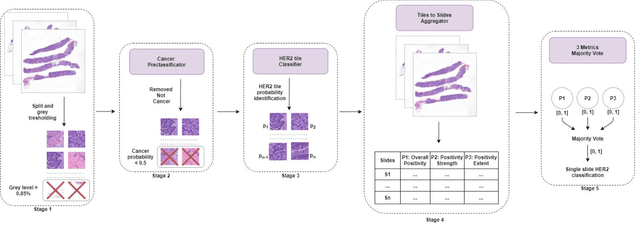
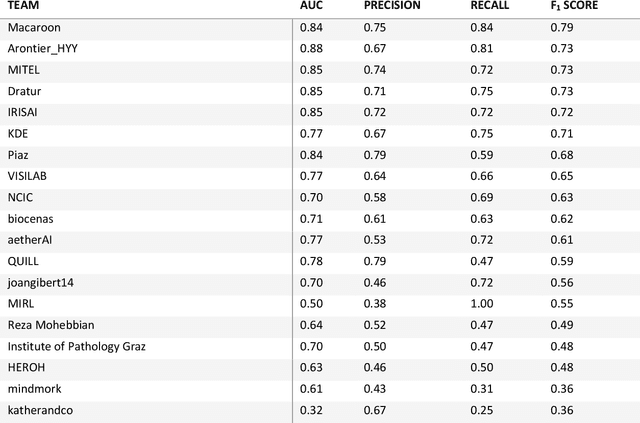
Abstract:Breast cancer is the most common malignancy in women, being responsible for more than half a million deaths every year. As such, early and accurate diagnosis is of paramount importance. Human expertise is required to diagnose and correctly classify breast cancer and define appropriate therapy, which depends on the evaluation of the expression of different biomarkers such as the transmembrane protein receptor HER2. This evaluation requires several steps, including special techniques such as immunohistochemistry or in situ hybridization to assess HER2 status. With the goal of reducing the number of steps and human bias in diagnosis, the HEROHE Challenge was organized, as a parallel event of the 16th European Congress on Digital Pathology, aiming to automate the assessment of the HER2 status based only on hematoxylin and eosin stained tissue sample of invasive breast cancer. Methods to assess HER2 status were presented by 21 teams worldwide and the results achieved by some of the proposed methods open potential perspectives to advance the state-of-the-art.
Certainty Pooling for Multiple Instance Learning
Aug 24, 2020
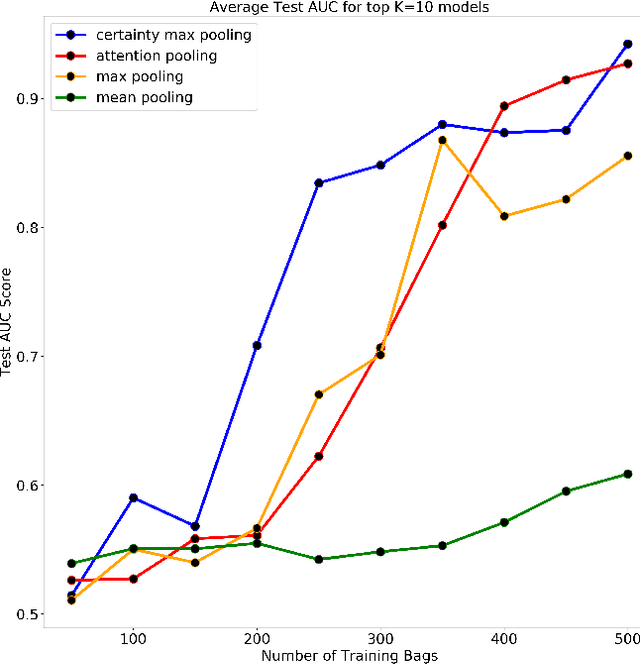
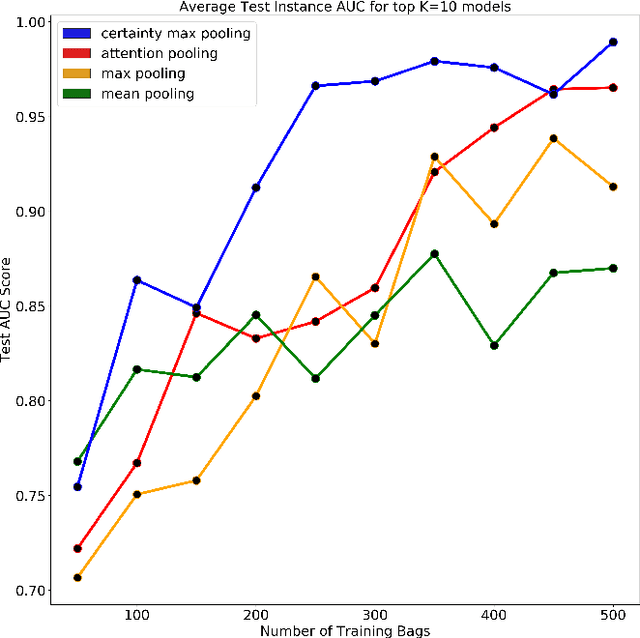
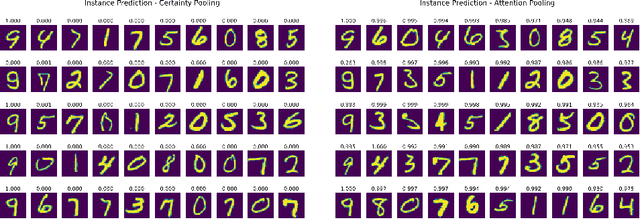
Abstract:Multiple Instance Learning is a form of weakly supervised learning in which the data is arranged in sets of instances called bags with one label assigned per bag. The bag level class prediction is derived from the multiple instances through application of a permutation invariant pooling operator on instance predictions or embeddings. We present a novel pooling operator called \textbf{Certainty Pooling} which incorporates the model certainty into bag predictions resulting in a more robust and explainable model. We compare our proposed method with other pooling operators in controlled experiments with low evidence ratio bags based on MNIST, as well as on a real life histopathology dataset - Camelyon16. Our method outperforms other methods in both bag level and instance level prediction, especially when only small training sets are available. We discuss the rationale behind our approach and the reasons for its superiority for these types of datasets.
Perceptual Embedding Consistency for Seamless Reconstruction of Tilewise Style Transfer
Jun 03, 2019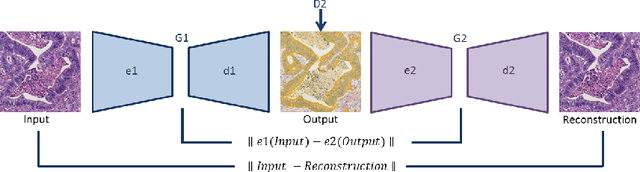

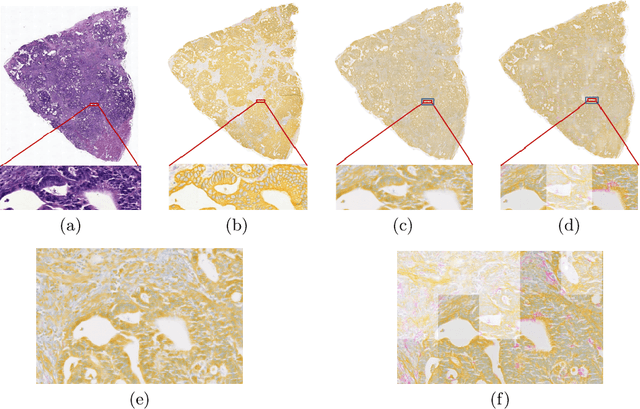
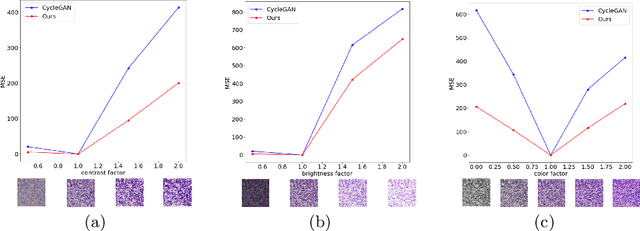
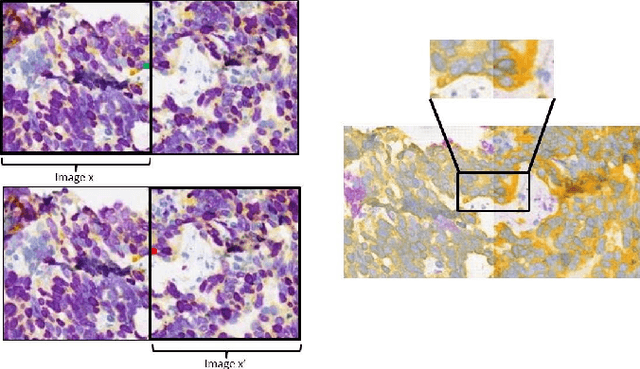
Abstract:Style transfer is a field with growing interest and use cases in deep learning. Recent work has shown Generative Adversarial Networks(GANs) can be used to create realistic images of virtually stained slide images in digital pathology with clinically validated interpretability. Digital pathology images are typically of extremely high resolution, making tilewise analysis necessary for deep learning applications. It has been shown that image generators with instance normalization can cause a tiling artifact when a large image is reconstructed from the tilewise analysis. We introduce a novel perceptual embedding consistency loss significantly reducing the tiling artifact created in the reconstructed whole slide image (WSI). We validate our results by comparing virtually stained slide images with consecutive real stained tissue slide images. We also demonstrate that our model is more robust to contrast, color and brightness perturbations by running comparative sensitivity analysis tests.
Self-Supervised Similarity Learning for Digital Pathology
May 20, 2019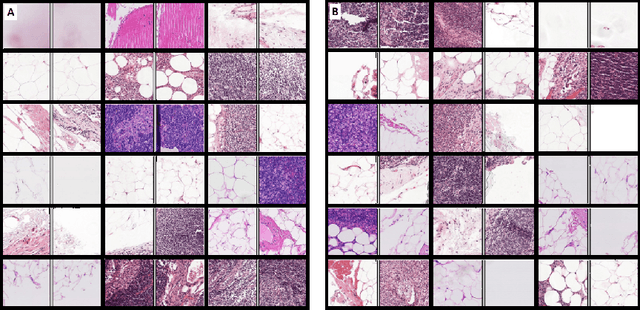

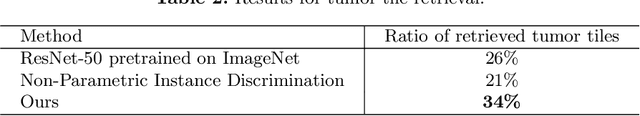
Abstract:Using features extracted from networks pretrained on ImageNet is a common practice in applications of deep learning for digital pathology. However it presents the downside of missing domain specific image information. In digital pathology, supervised training data is expensive and difficult to collect. We propose a self supervised method for feature extraction by similarity learning on whole slide images (WSI) that is simple to implement and allows creation of robust and compact image descriptors. We train a siamese network, exploiting image spatial continuity and assuming spatially adjacent tiles in the image are more similar to each other than distant tiles. Our network outputs feature vectors of length 128, which allows dramatically lower memory storage and faster processing than networks pretrained on ImageNet. We apply the method on digital pathology whole slide images (WSI) from the Camelyon16 train set and assess and compare our method by measuring image retrieval of tumor tiles and descriptor pair distance ratio for distant/near tiles in the Camelyon16 test set. We show that our method yields better retrieval task results than existing ImageNet based and generic self-supervised feature extraction methods. To the best of our knowledge, this is also the first published method for self supervised learning tailored for digital pathology.
Virtualization of tissue staining in digital pathology using an unsupervised deep learning approach
Oct 15, 2018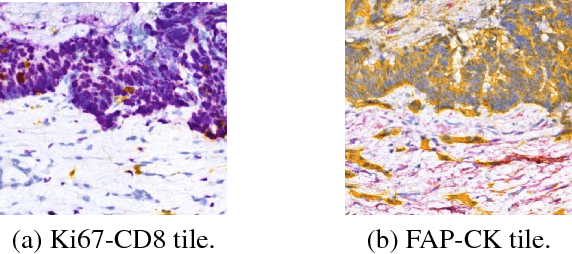

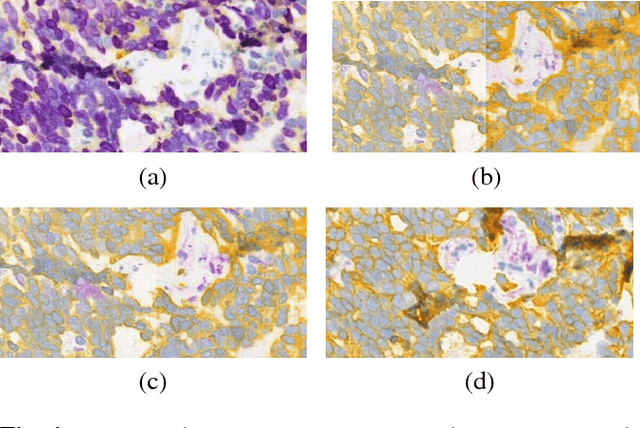
Abstract:Histopathological evaluation of tissue samples is a key practice in patient diagnosis and drug development, especially in oncology. Historically, Hematoxylin and Eosin (H&E) has been used by pathologists as a gold standard staining. However, in many cases, various target specific stains, including immunohistochemistry (IHC), are needed in order to highlight specific structures in the tissue. As tissue is scarce and staining procedures are tedious, it would be beneficial to generate images of stained tissue virtually. Virtual staining could also generate in-silico multiplexing of different stains on the same tissue segment. In this paper, we present a sample application that generates FAP-CK virtual IHC images from Ki67-CD8 real IHC images using an unsupervised deep learning approach based on CycleGAN. We also propose a method to deal with tiling artifacts caused by normalization layers and we validate our approach by comparing the results of tissue analysis algorithms for virtual and real images.
Generalizing multistain immunohistochemistry tissue segmentation using one-shot color deconvolution deep neural networks
Sep 22, 2018

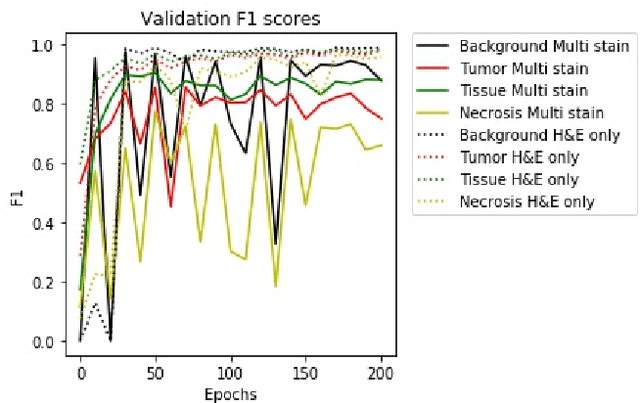
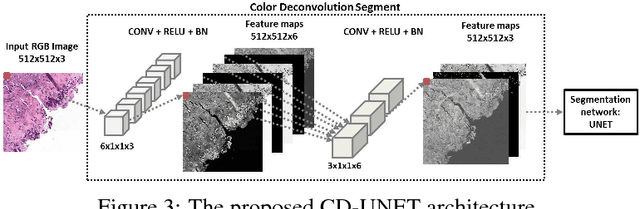
Abstract:A key challenge in cancer immunotherapy biomarker research is quantification of pattern changes in microscopic whole slide images of tumor biopsies. Different cell types tend to migrate into various tissue compartments and form variable distribution patterns. Drug development requires correlative analysis of various biomarkers in and between the tissue compartments. To enable that, tissue slides are manually annotated by expert pathologists. Manual annotation of tissue slides is a labor intensive, tedious and error-prone task. Automation of this annotation process can improve accuracy and consistency while reducing workload and cost in a way that will positively influence drug development efforts. In this paper we present a novel one-shot color deconvolution deep learning method to automatically segment and annotate digitized slide images with multiple stainings into compartments of tumor, healthy tissue, and necrosis. We address the task in the context of drug development where multiple stains, tissue and tumor types exist and look into solutions for generalizations over these image populations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge