Ekaterina Kondrateva
Benchmarking the Reproducibility of Brain MRI Segmentation Across Scanners and Time
Apr 22, 2025



Abstract:Accurate and reproducible brain morphometry from structural MRI is critical for monitoring neuroanatomical changes across time and across imaging domains. Although deep learning has accelerated segmentation workflows, scanner-induced variability and reproducibility limitations remain-especially in longitudinal and multi-site settings. In this study, we benchmark two modern segmentation pipelines, FastSurfer and SynthSeg, both integrated into FreeSurfer, one of the most widely adopted tools in neuroimaging. Using two complementary datasets - a 17-year longitudinal cohort (SIMON) and a 9-site test-retest cohort (SRPBS)-we quantify inter-scan segmentation variability using Dice coefficient, Surface Dice, Hausdorff Distance (HD95), and Mean Absolute Percentage Error (MAPE). Our results reveal up to 7-8% volume variation in small subcortical structures such as the amygdala and ventral diencephalon, even under controlled test-retest conditions. This raises a key question: is it feasible to detect subtle longitudinal changes on the order of 5-10% in pea-sized brain regions, given the magnitude of domain-induced morphometric noise? We further analyze the effects of registration templates and interpolation modes, and propose surface-based quality filtering to improve segmentation reliability. This study provides a reproducible benchmark for morphometric reproducibility and emphasizes the need for harmonization strategies in real-world neuroimaging studies. Code and figures: https://github.com/kondratevakate/brain-mri-segmentation
Neglectable effect of brain MRI data prepreprocessing for tumor segmentation
Apr 11, 2022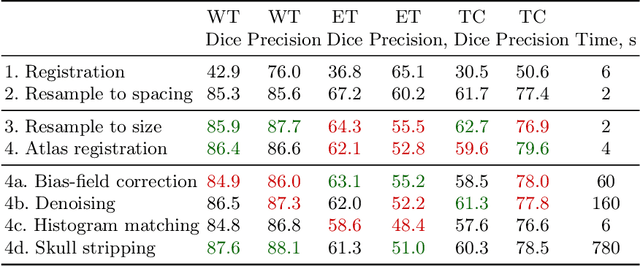
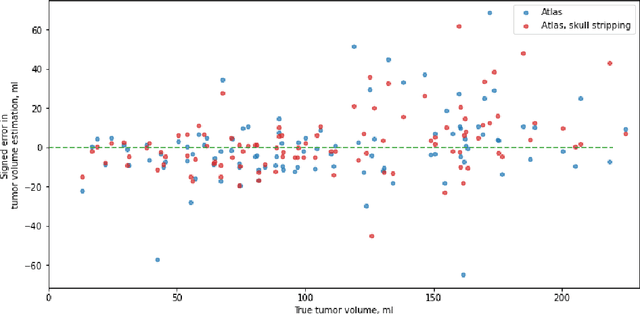
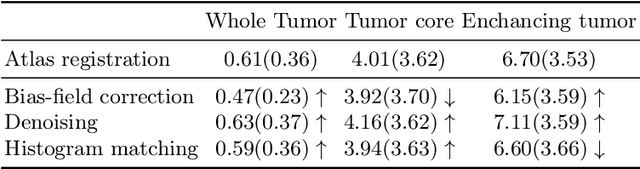
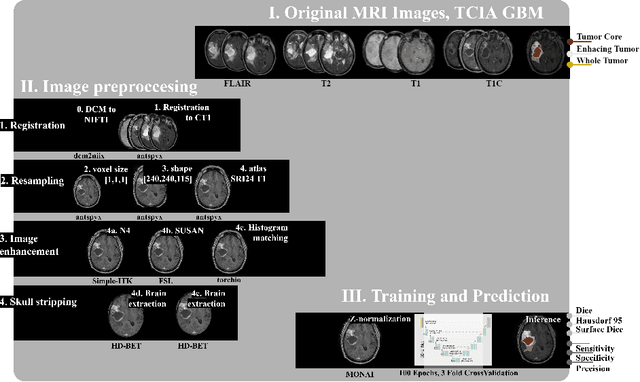
Abstract:Magnetic resonance imaging (MRI) data is heterogeneous due to the differences in device manufacturers, scanning protocols, and inter-subject variability. A conventional way to mitigate MR image heterogeneity is to apply preprocessing transformations, such as anatomy alignment, voxel resampling, signal intensity equalization, image denoising, and localization of regions of interest (ROI). Although preprocessing pipeline standardizes image appearance, its influence on the quality of image segmentation and other downstream tasks on deep neural networks (DNN) has never been rigorously studied. Here we report a comprehensive study of multimodal MRI brain cancer image segmentation on TCIA-GBM open-source dataset. Our results demonstrate that most popular standardization steps add no value to artificial neural network performance; moreover, preprocessing can hamper model performance. We suggest that image intensity normalization approaches do not contribute to model accuracy because of the reduction of signal variance with image standardization. Finally, we show the contribution of scull-stripping in data preprocessing is almost negligible if measured in terms of clinically relevant metrics. We show that the only essential transformation for accurate analysis is the unification of voxel spacing across the dataset. In contrast, anatomy alignment in form of non-rigid atlas registration is not necessary and most intensity equalization steps do not improve model productiveness.
Convolutional neural networks for automatic detection of Focal Cortical Dysplasia
Oct 20, 2020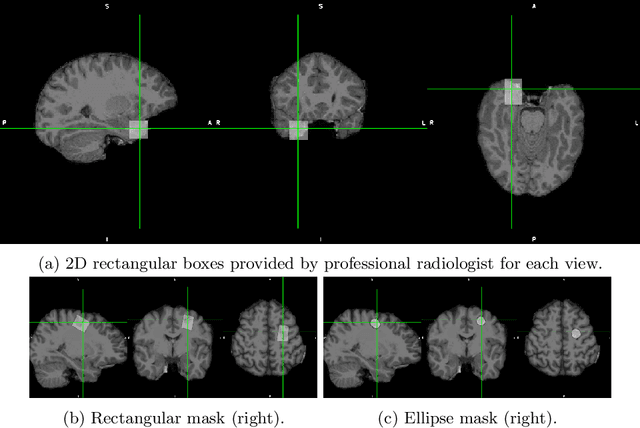
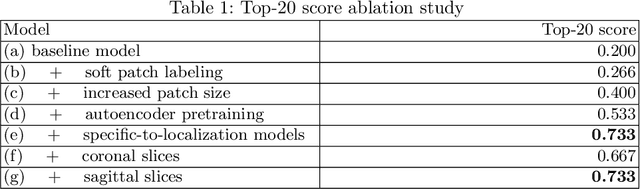
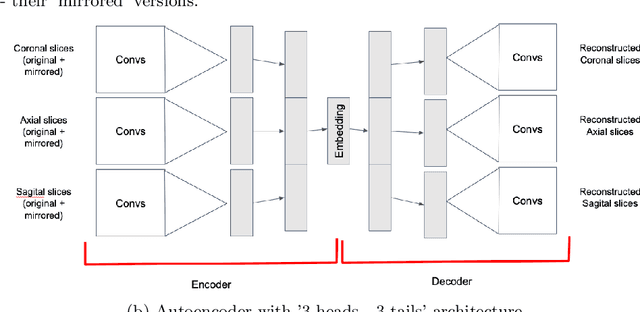
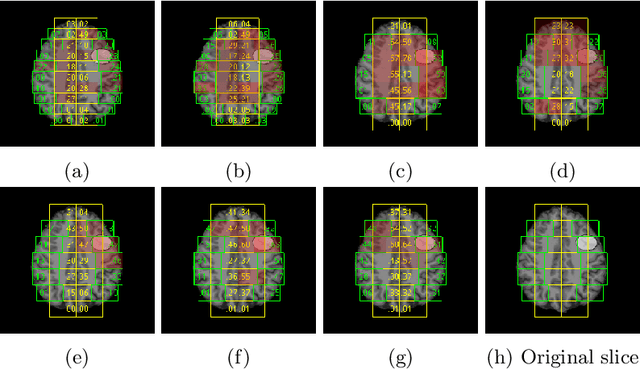
Abstract:Focal cortical dysplasia (FCD) is one of the most common epileptogenic lesions associated with cortical development malformations. However, the accurate detection of the FCD relies on the radiologist professionalism, and in many cases, the lesion could be missed. In this work, we solve the problem of automatic identification of FCD on magnetic resonance images (MRI). For this task, we improve recent methods of Deep Learning-based FCD detection and apply it for a dataset of 15 labeled FCD patients. The model results in the successful detection of FCD on 11 out of 15 subjects.
Fader Networks for domain adaptation on fMRI: ABIDE-II study
Oct 14, 2020Abstract:ABIDE is the largest open-source autism spectrum disorder database with both fMRI data and full phenotype description. These data were extensively studied based on functional connectivity analysis as well as with deep learning on raw data, with top models accuracy close to 75\% for separate scanning sites. Yet there is still a problem of models transferability between different scanning sites within ABIDE. In the current paper, we for the first time perform domain adaptation for brain pathology classification problem on raw neuroimaging data. We use 3D convolutional autoencoders to build the domain irrelevant latent space image representation and demonstrate this method to outperform existing approaches on ABIDE data.
Domain Shift in Computer Vision models for MRI data analysis: An Overview
Oct 14, 2020Abstract:Machine learning and computer vision methods are showing good performance in medical imagery analysis. Yetonly a few applications are now in clinical use and one of the reasons for that is poor transferability of themodels to data from different sources or acquisition domains. Development of new methods and algorithms forthe transfer of training and adaptation of the domain in multi-modal medical imaging data is crucial for thedevelopment of accurate models and their use in clinics. In present work, we overview methods used to tackle thedomain shift problem in machine learning and computer vision. The algorithms discussed in this survey includeadvanced data processing, model architecture enhancing and featured training, as well as predicting in domaininvariant latent space. The application of the autoencoding neural networks and their domain-invariant variationsare heavily discussed in a survey. We observe the latest methods applied to the magnetic resonance imaging(MRI) data analysis and conclude on their performance as well as propose directions for further research.
* 8 pages, 1 figure
Interpretable Deep Learning for Pattern Recognition in Brain Differences Between Men and Women
Jun 20, 2020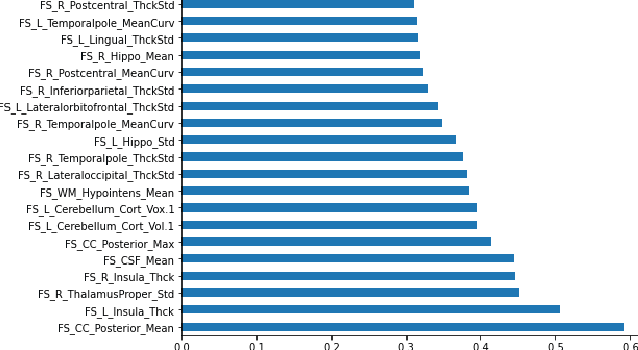
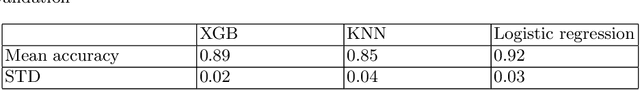
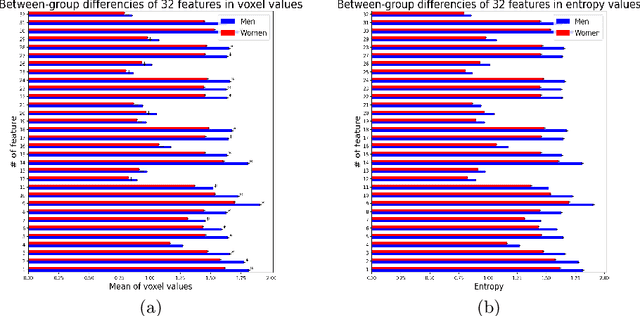
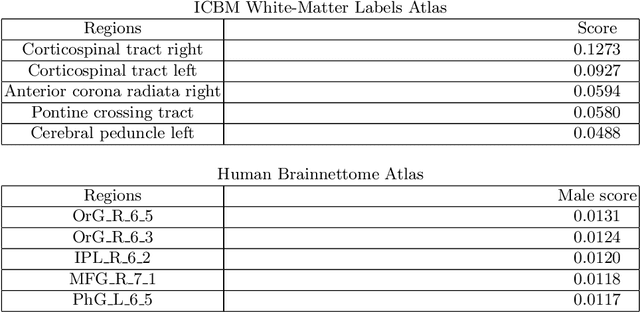
Abstract:Deep learning shows high potential for many medical image analysis tasks. Neural networks work with full-size data without extensive preprocessing and feature generation and, thus, information loss. Recent work has shown that morphological difference between specific brain regions can be found on MRI with deep learning techniques. We consider the pattern recognition task based on a large open-access dataset of healthy subjects - an exploration of brain differences between men and women. However, interpretation of the lately proposed models is based on a region of interest and can not be extended to pixel or voxel-wise image interpretation, which is considered to be more informative. In this paper, we confirm the previous findings in sex differences from diffusion-tensor imaging on T1 weighted brain MRI scans. We compare the results of three voxel-based 3D CNN interpretation methods: Meaningful Perturbations, GradCam and Guided Backpropagation and provide the open-source code.
3D Deformable Convolutions for MRI classification
Nov 05, 2019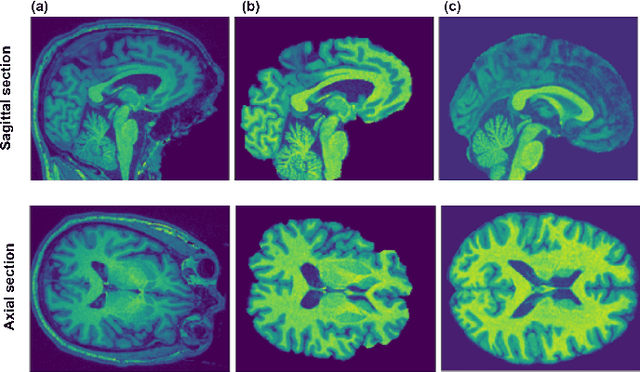
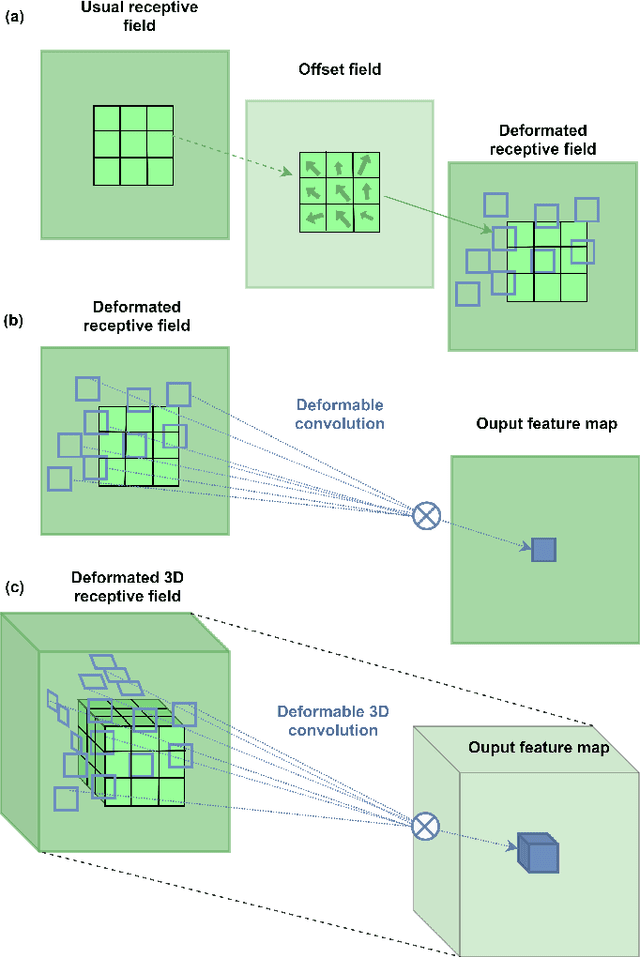
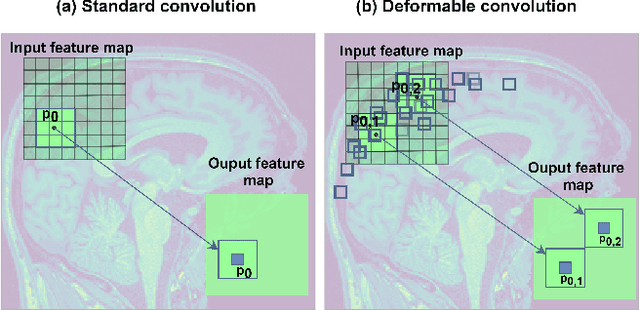
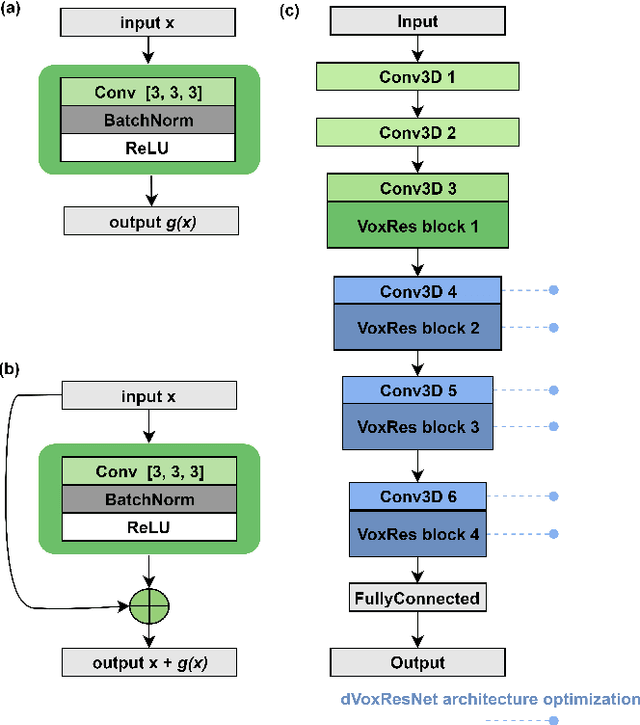
Abstract:Deep learning convolutional neural networks have proved to be a powerful tool for MRI analysis. In current work, we explore the potential of the deformable convolutional deep neural network layers for MRI data classification. We propose new 3D deformable convolutions(d-convolutions), implement them in VoxResNet architecture and apply for structural MRI data classification. We show that 3D d-convolutions outperform standard ones and are effective for unprocessed 3D MR images being robust to particular geometrical properties of the data. Firstly proposed dVoxResNet architecture exhibits high potential for the use in MRI data classification.
Ensemble of 3D CNN regressors with data fusion for fluid intelligence prediction
May 25, 2019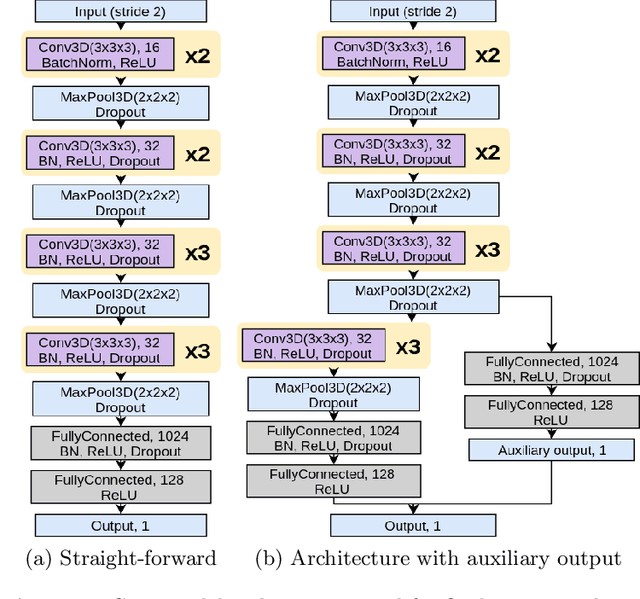
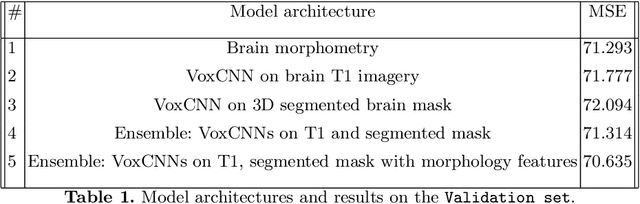
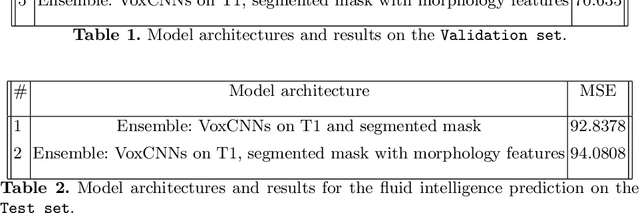
Abstract:In this work, we aim at predicting children's fluid intelligence scores based on structural T1-weighted MR images from the largest long-term study of brain development and child health. The target variable was regressed on a data collection site, socio-demographic variables and brain volume, thus being independent to the potentially informative factors, which are not directly related to the brain functioning. We investigate both feature extraction and deep learning approaches as well as different deep CNN architectures and their ensembles. We propose an advanced architecture of VoxCNNs ensemble, which yield MSE (92.838) on blind test.
* 10 pages, 1 figure, 2 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge