Alexander Bernstein
Multivariate Wasserstein Functional Connectivity for Autism Screening
Sep 23, 2022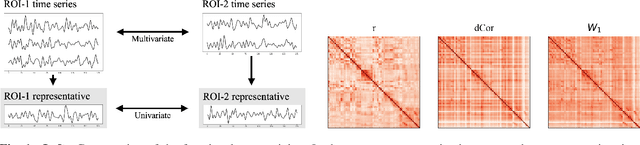
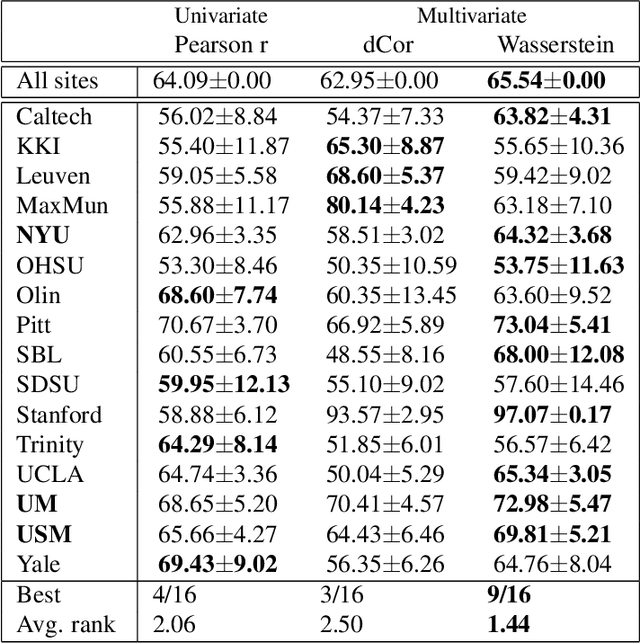
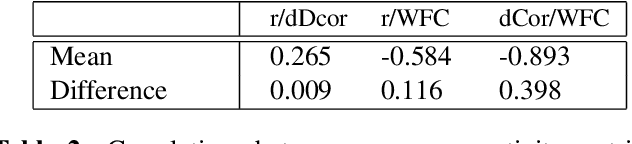
Abstract:Most approaches to the estimation of brain functional connectivity from the functional magnetic resonance imaging (fMRI) data rely on computing some measure of statistical dependence, or more generally, a distance between univariate representative time series of regions of interest (ROIs) consisting of multiple voxels. However, summarizing a ROI's multiple time series with its mean or the first principal component (1PC) may result to the loss of information as, for example, 1PC explains only a small fraction of variance of the multivariate signal of the neuronal activity. We propose to compare ROIs directly, without the use of representative time series, defining a new measure of multivariate connectivity between ROIs, not necessarily consisting of the same number of voxels, based on the Wasserstein distance. We assess the proposed Wasserstein functional connectivity measure on the autism screening task, demonstrating its superiority over commonly used univariate and multivariate functional connectivity measures.
Artificial Text Detection via Examining the Topology of Attention Maps
Sep 10, 2021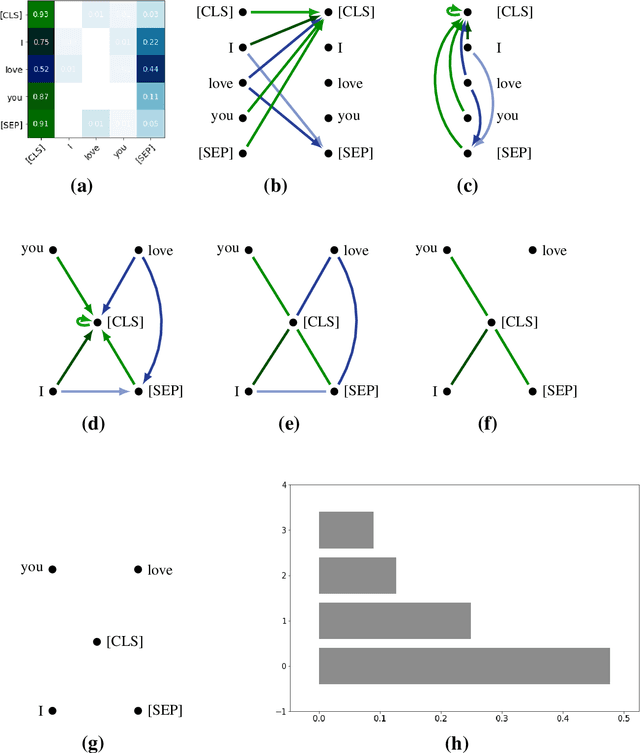
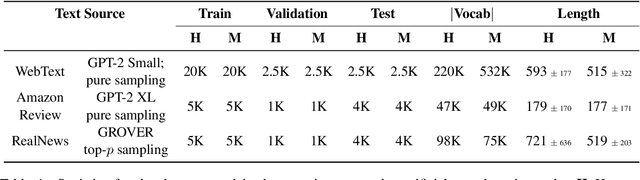
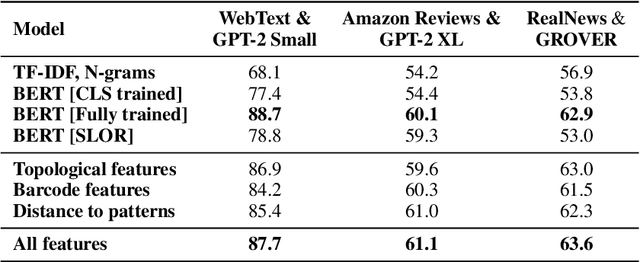
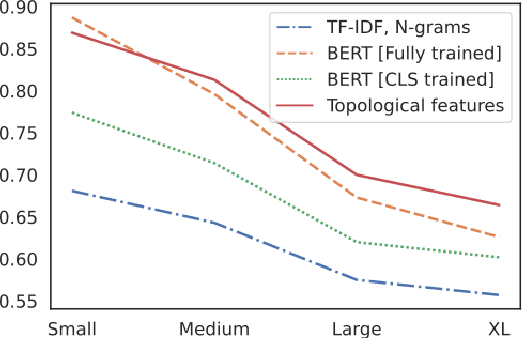
Abstract:The impressive capabilities of recent generative models to create texts that are challenging to distinguish from the human-written ones can be misused for generating fake news, product reviews, and even abusive content. Despite the prominent performance of existing methods for artificial text detection, they still lack interpretability and robustness towards unseen models. To this end, we propose three novel types of interpretable topological features for this task based on Topological Data Analysis (TDA) which is currently understudied in the field of NLP. We empirically show that the features derived from the BERT model outperform count- and neural-based baselines up to 10\% on three common datasets, and tend to be the most robust towards unseen GPT-style generation models as opposed to existing methods. The probing analysis of the features reveals their sensitivity to the surface and syntactic properties. The results demonstrate that TDA is a promising line with respect to NLP tasks, specifically the ones that incorporate surface and structural information.
Convolutional neural networks for automatic detection of Focal Cortical Dysplasia
Oct 20, 2020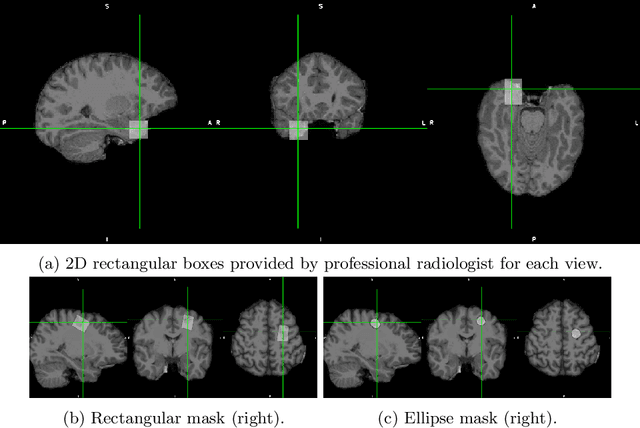
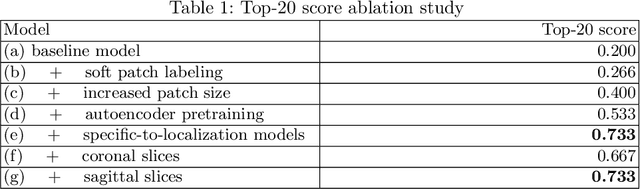
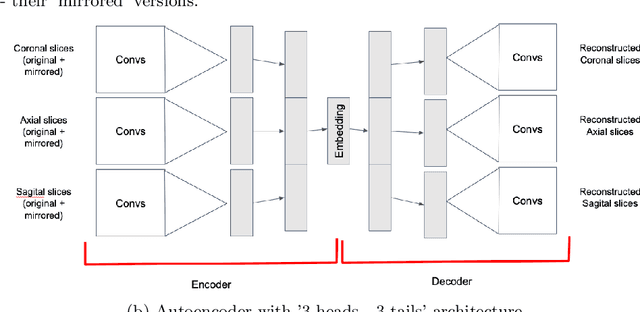
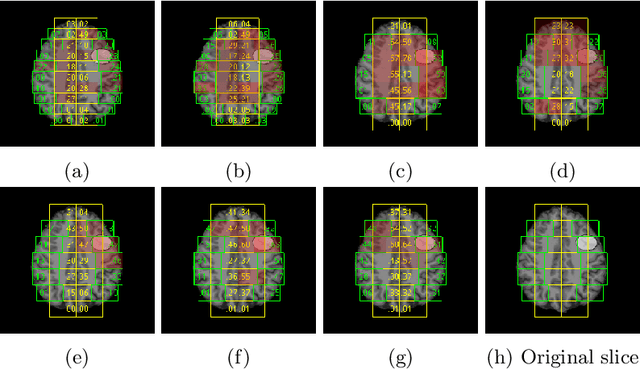
Abstract:Focal cortical dysplasia (FCD) is one of the most common epileptogenic lesions associated with cortical development malformations. However, the accurate detection of the FCD relies on the radiologist professionalism, and in many cases, the lesion could be missed. In this work, we solve the problem of automatic identification of FCD on magnetic resonance images (MRI). For this task, we improve recent methods of Deep Learning-based FCD detection and apply it for a dataset of 15 labeled FCD patients. The model results in the successful detection of FCD on 11 out of 15 subjects.
Fader Networks for domain adaptation on fMRI: ABIDE-II study
Oct 14, 2020Abstract:ABIDE is the largest open-source autism spectrum disorder database with both fMRI data and full phenotype description. These data were extensively studied based on functional connectivity analysis as well as with deep learning on raw data, with top models accuracy close to 75\% for separate scanning sites. Yet there is still a problem of models transferability between different scanning sites within ABIDE. In the current paper, we for the first time perform domain adaptation for brain pathology classification problem on raw neuroimaging data. We use 3D convolutional autoencoders to build the domain irrelevant latent space image representation and demonstrate this method to outperform existing approaches on ABIDE data.
Domain Shift in Computer Vision models for MRI data analysis: An Overview
Oct 14, 2020Abstract:Machine learning and computer vision methods are showing good performance in medical imagery analysis. Yetonly a few applications are now in clinical use and one of the reasons for that is poor transferability of themodels to data from different sources or acquisition domains. Development of new methods and algorithms forthe transfer of training and adaptation of the domain in multi-modal medical imaging data is crucial for thedevelopment of accurate models and their use in clinics. In present work, we overview methods used to tackle thedomain shift problem in machine learning and computer vision. The algorithms discussed in this survey includeadvanced data processing, model architecture enhancing and featured training, as well as predicting in domaininvariant latent space. The application of the autoencoding neural networks and their domain-invariant variationsare heavily discussed in a survey. We observe the latest methods applied to the magnetic resonance imaging(MRI) data analysis and conclude on their performance as well as propose directions for further research.
* 8 pages, 1 figure
Interpretable Deep Learning for Pattern Recognition in Brain Differences Between Men and Women
Jun 20, 2020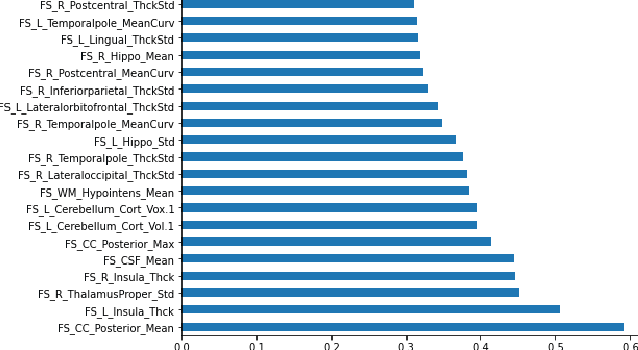
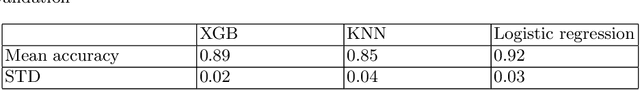
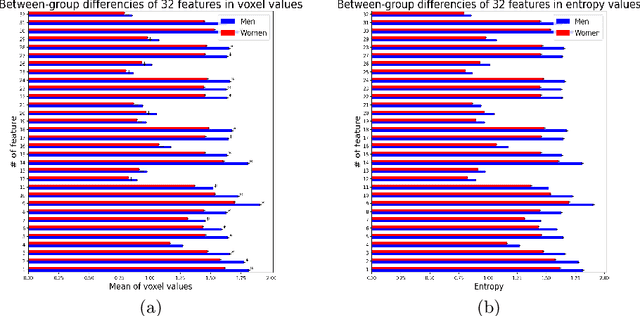
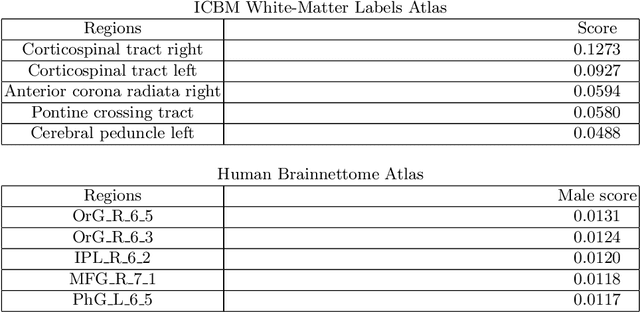
Abstract:Deep learning shows high potential for many medical image analysis tasks. Neural networks work with full-size data without extensive preprocessing and feature generation and, thus, information loss. Recent work has shown that morphological difference between specific brain regions can be found on MRI with deep learning techniques. We consider the pattern recognition task based on a large open-access dataset of healthy subjects - an exploration of brain differences between men and women. However, interpretation of the lately proposed models is based on a region of interest and can not be extended to pixel or voxel-wise image interpretation, which is considered to be more informative. In this paper, we confirm the previous findings in sex differences from diffusion-tensor imaging on T1 weighted brain MRI scans. We compare the results of three voxel-based 3D CNN interpretation methods: Meaningful Perturbations, GradCam and Guided Backpropagation and provide the open-source code.
Weakly Supervised Fine Tuning Approach for Brain Tumor Segmentation Problem
Nov 06, 2019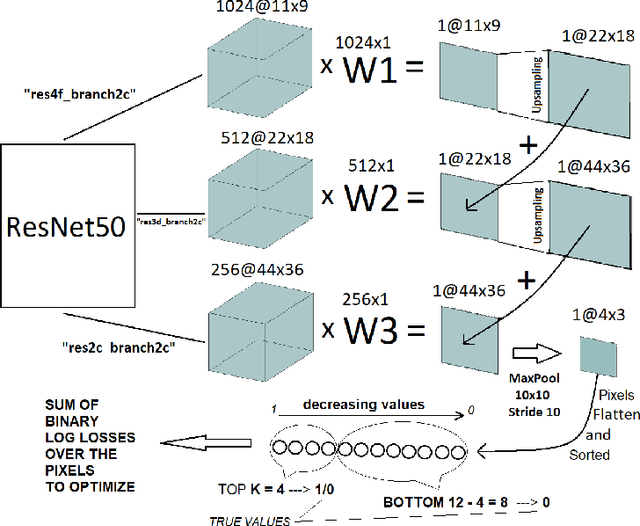
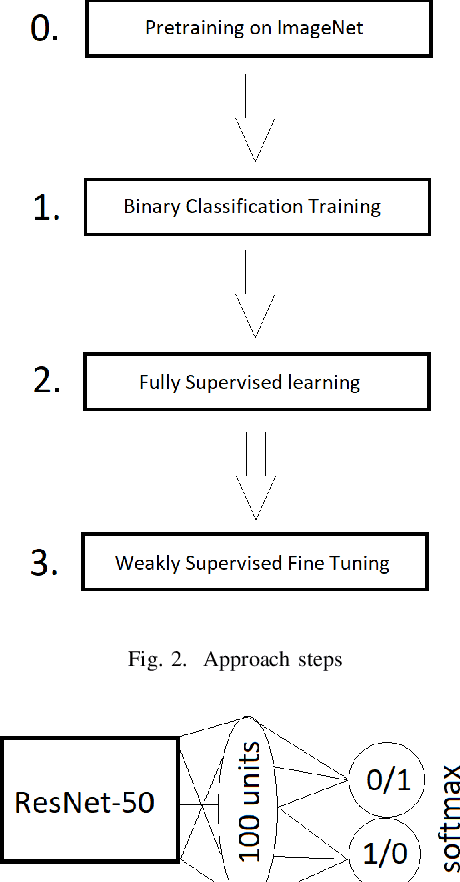
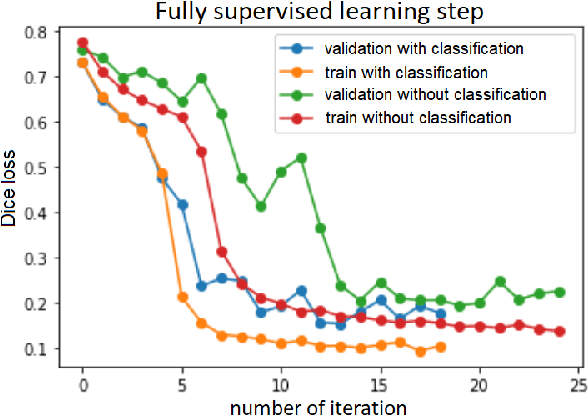
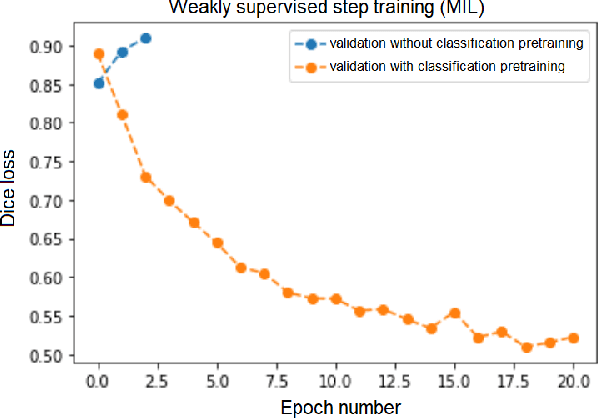
Abstract:Segmentation of tumors in brain MRI images is a challenging task, where most recent methods demand large volumes of data with pixel-level annotations, which are generally costly to obtain. In contrast, image-level annotations, where only the presence of lesion is marked, are generally cheap, generated in far larger volumes compared to pixel-level labels, and contain less labeling noise. In the context of brain tumor segmentation, both pixel-level and image-level annotations are commonly available; thus, a natural question arises whether a segmentation procedure could take advantage of both. In the present work we: 1) propose a learning-based framework that allows simultaneous usage of both pixel- and image-level annotations in MRI images to learn a segmentation model for brain tumor; 2) study the influence of comparative amounts of pixel- and image-level annotations on the quality of brain tumor segmentation; 3) compare our approach to the traditional fully-supervised approach and show that the performance of our method in terms of segmentation quality may be competitive.
3D Deformable Convolutions for MRI classification
Nov 05, 2019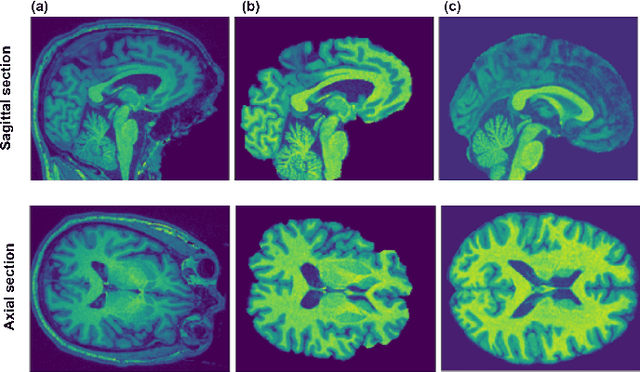
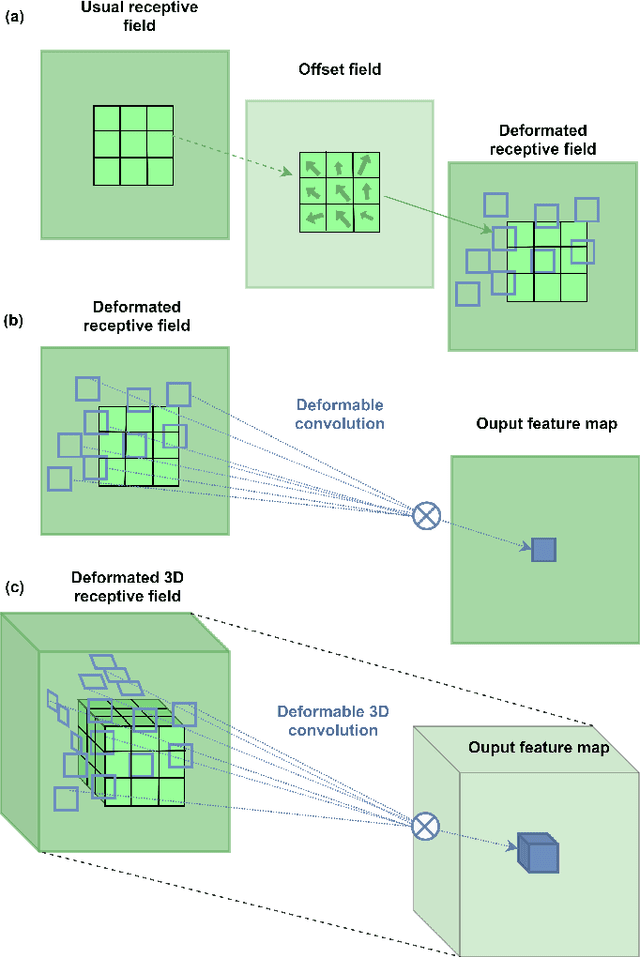
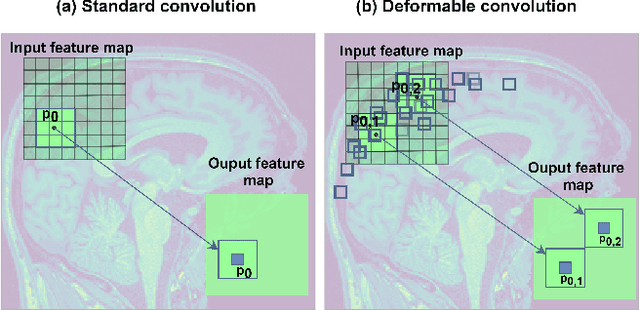
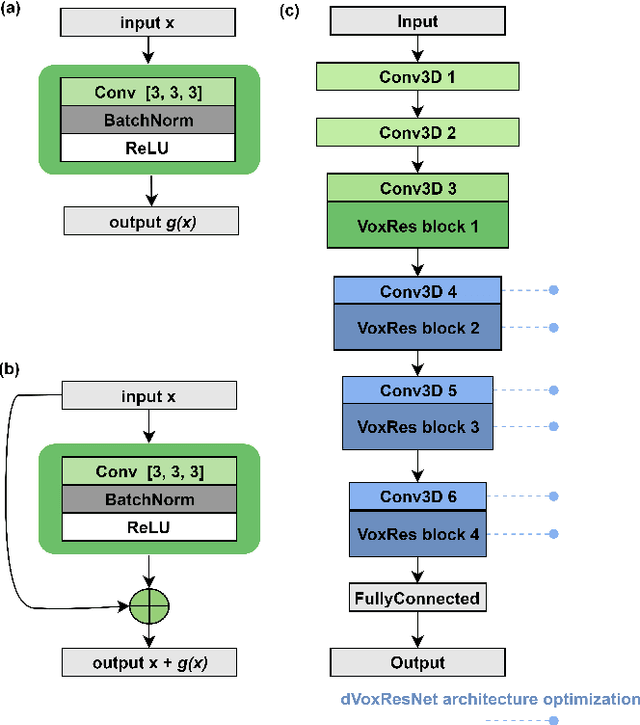
Abstract:Deep learning convolutional neural networks have proved to be a powerful tool for MRI analysis. In current work, we explore the potential of the deformable convolutional deep neural network layers for MRI data classification. We propose new 3D deformable convolutions(d-convolutions), implement them in VoxResNet architecture and apply for structural MRI data classification. We show that 3D d-convolutions outperform standard ones and are effective for unprocessed 3D MR images being robust to particular geometrical properties of the data. Firstly proposed dVoxResNet architecture exhibits high potential for the use in MRI data classification.
fMRI: preprocessing, classification and pattern recognition
Apr 26, 2018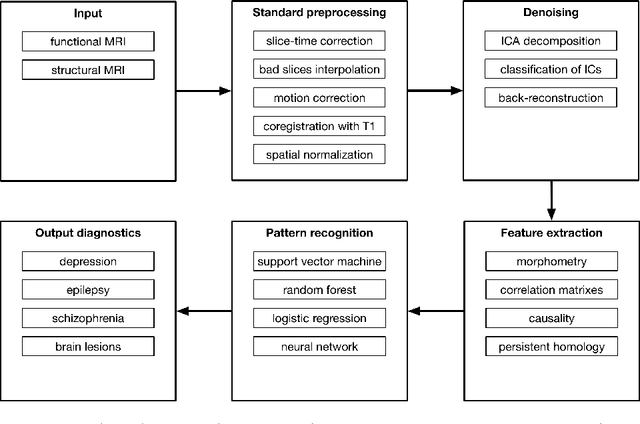

Abstract:As machine learning continues to gain momentum in the neuroscience community, we witness the emergence of novel applications such as diagnostics, characterization, and treatment outcome prediction for psychiatric and neurological disorders, for instance, epilepsy and depression. Systematic research into these mental disorders increasingly involves drawing clinical conclusions on the basis of data-driven approaches; to this end, structural and functional neuroimaging serve as key source modalities. Identification of informative neuroimaging markers requires establishing a comprehensive preparation pipeline for data which may be severely corrupted by artifactual signal fluctuations. In this work, we review a large body of literature to provide ample evidence for the advantages of pattern recognition approaches in clinical applications, overview advanced graph-based pattern recognition approaches, and propose a noise-aware neuroimaging data processing pipeline. To demonstrate the effectiveness of our approach, we provide results from a pilot study, which show a significant improvement in classification accuracy, indicating a promising research direction.
Machine Learning pipeline for discovering neuroimaging-based biomarkers in neurology and psychiatry
Apr 26, 2018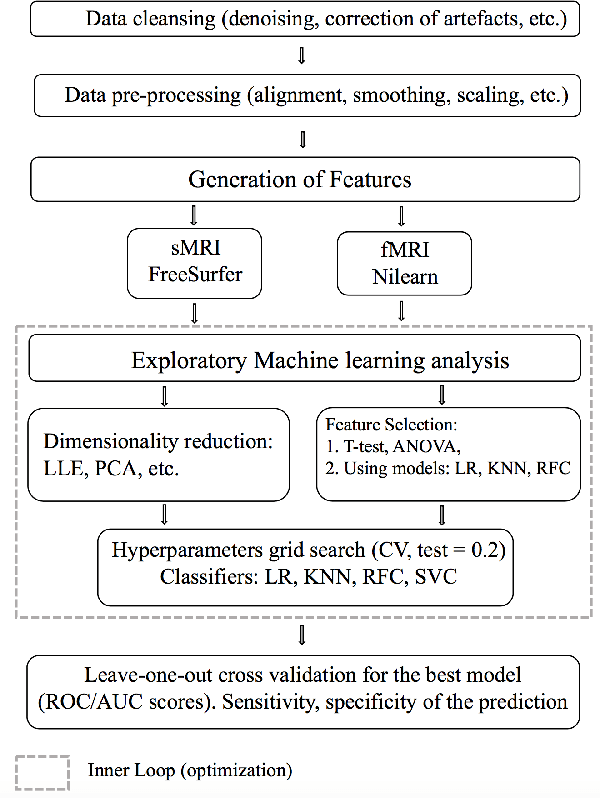
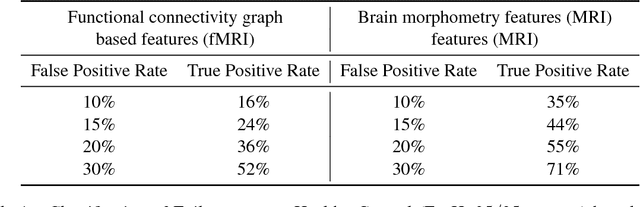
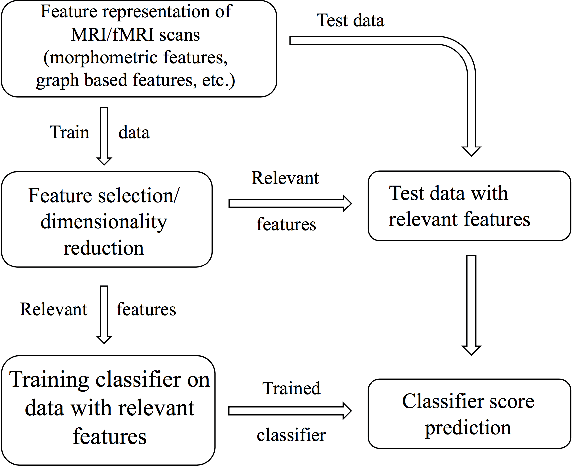
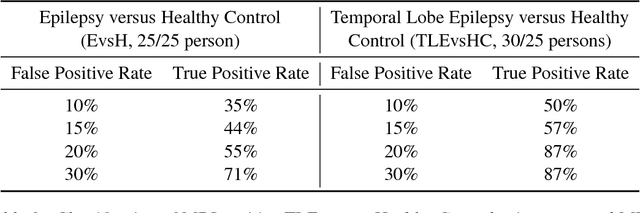
Abstract:We consider a problem of diagnostic pattern recognition/classification from neuroimaging data. We propose a common data analysis pipeline for neuroimaging-based diagnostic classification problems using various ML algorithms and processing toolboxes for brain imaging. We illustrate the pipeline application by discovering new biomarkers for diagnostics of epilepsy and depression based on clinical and MRI/fMRI data for patients and healthy volunteers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge