David E. Carlson
Pose Splatter: A 3D Gaussian Splatting Model for Quantifying Animal Pose and Appearance
May 23, 2025Abstract:Accurate and scalable quantification of animal pose and appearance is crucial for studying behavior. Current 3D pose estimation techniques, such as keypoint- and mesh-based techniques, often face challenges including limited representational detail, labor-intensive annotation requirements, and expensive per-frame optimization. These limitations hinder the study of subtle movements and can make large-scale analyses impractical. We propose Pose Splatter, a novel framework leveraging shape carving and 3D Gaussian splatting to model the complete pose and appearance of laboratory animals without prior knowledge of animal geometry, per-frame optimization, or manual annotations. We also propose a novel rotation-invariant visual embedding technique for encoding pose and appearance, designed to be a plug-in replacement for 3D keypoint data in downstream behavioral analyses. Experiments on datasets of mice, rats, and zebra finches show Pose Splatter learns accurate 3D animal geometries. Notably, Pose Splatter represents subtle variations in pose, provides better low-dimensional pose embeddings over state-of-the-art as evaluated by humans, and generalizes to unseen data. By eliminating annotation and per-frame optimization bottlenecks, Pose Splatter enables analysis of large-scale, longitudinal behavior needed to map genotype, neural activity, and micro-behavior at unprecedented resolution.
Distillation Learning Guided by Image Reconstruction for One-Shot Medical Image Segmentation
Aug 07, 2024



Abstract:Traditional one-shot medical image segmentation (MIS) methods use registration networks to propagate labels from a reference atlas or rely on comprehensive sampling strategies to generate synthetic labeled data for training. However, these methods often struggle with registration errors and low-quality synthetic images, leading to poor performance and generalization. To overcome this, we introduce a novel one-shot MIS framework based on knowledge distillation, which allows the network to directly 'see' real images through a distillation process guided by image reconstruction. It focuses on anatomical structures in a single labeled image and a few unlabeled ones. A registration-based data augmentation network creates realistic, labeled samples, while a feature distillation module helps the student network learn segmentation from these samples, guided by the teacher network. During inference, the streamlined student network accurately segments new images. Evaluations on three public datasets (OASIS for T1 brain MRI, BCV for abdomen CT, and VerSe for vertebrae CT) show superior segmentation performance and generalization across different medical image datasets and modalities compared to leading methods. Our code is available at https://github.com/NoviceFodder/OS-MedSeg.
On Target Shift in Adversarial Domain Adaptation
Mar 15, 2019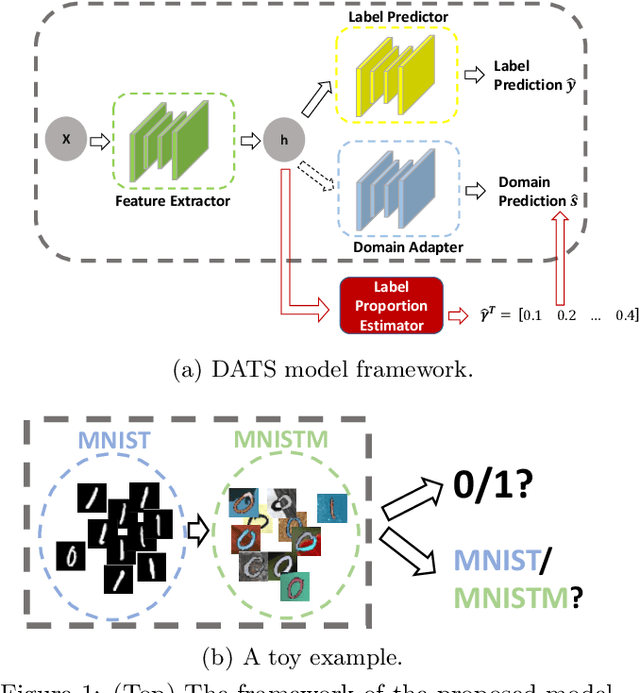
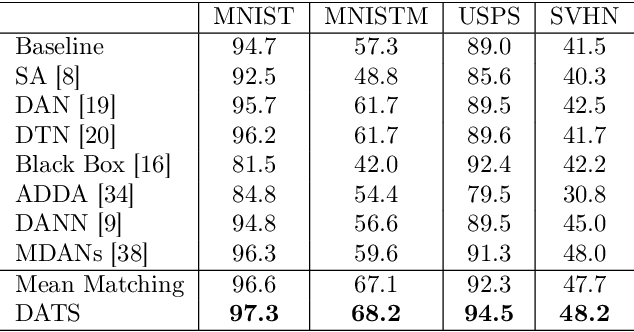
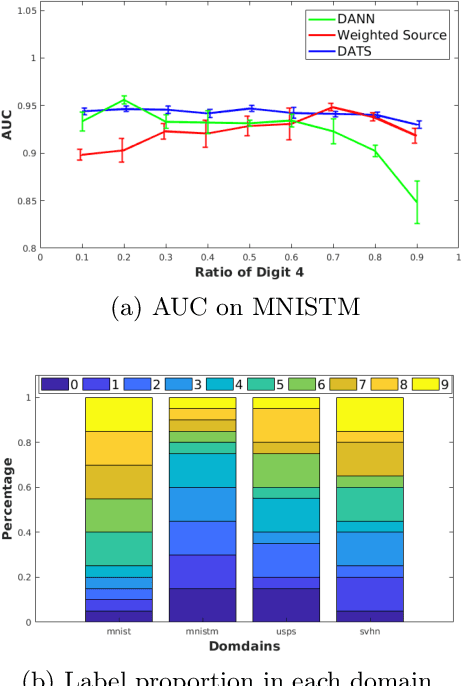
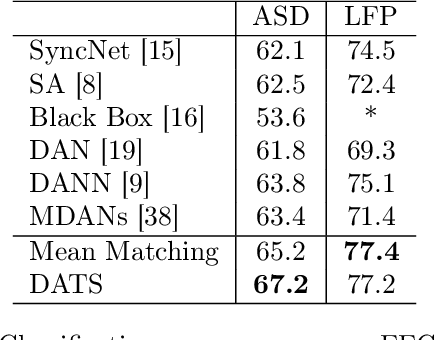
Abstract:Discrepancy between training and testing domains is a fundamental problem in the generalization of machine learning techniques. Recently, several approaches have been proposed to learn domain invariant feature representations through adversarial deep learning. However, label shift, where the percentage of data in each class is different between domains, has received less attention. Label shift naturally arises in many contexts, especially in behavioral studies where the behaviors are freely chosen. In this work, we propose a method called Domain Adversarial nets for Target Shift (DATS) to address label shift while learning a domain invariant representation. This is accomplished by using distribution matching to estimate label proportions in a blind test set. We extend this framework to handle multiple domains by developing a scheme to upweight source domains most similar to the target domain. Empirical results show that this framework performs well under large label shift in synthetic and real experiments, demonstrating the practical importance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge